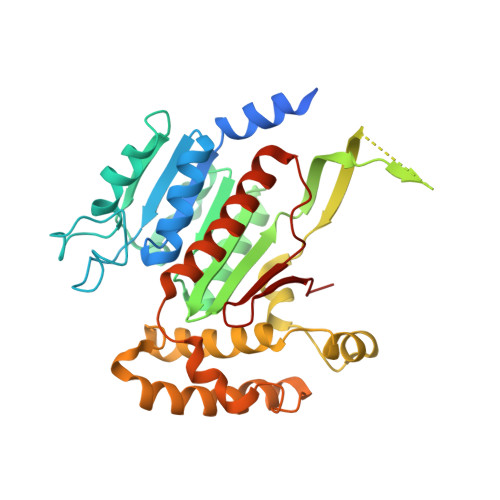
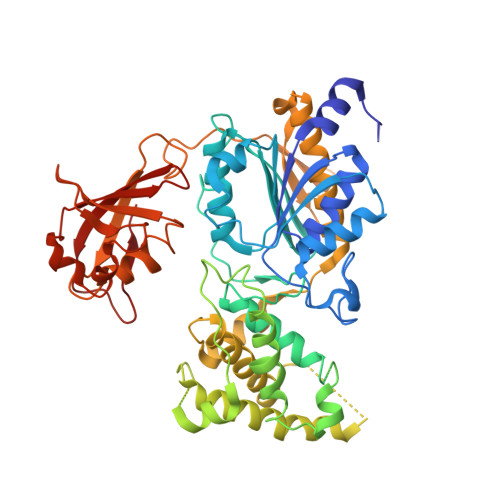
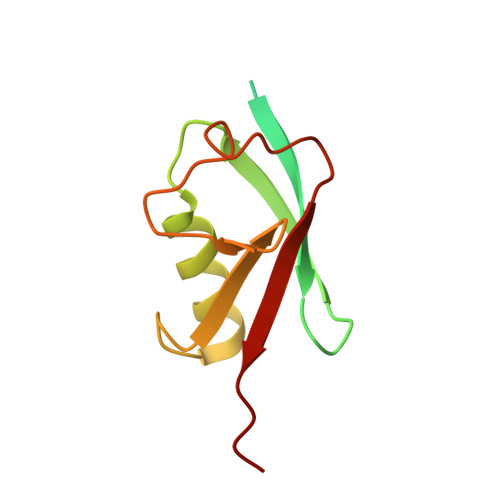
Active site remodelling accompanies thioester bond formation in the SUMO E1.
Olsen, S.K., Capili, A.D., Lu, X., Tan, D.S., Lima, C.D.(2010) Nature 463: 906-912
- PubMed: 20164921
- DOI: https://doi.org/10.1038/nature08765
- Primary Citation of Related Structures:
3KYC, 3KYD - PubMed Abstract:
E1 enzymes activate ubiquitin (Ub) and ubiquitin-like (Ubl) proteins in two steps by carboxy-terminal adenylation and thioester bond formation to a conserved catalytic cysteine in the E1 Cys domain. The structural basis for these intermediates remains unknown. Here we report crystal structures for human SUMO E1 in complex with SUMO adenylate and tetrahedral intermediate analogues at 2.45 and 2.6 A, respectively. These structures show that side chain contacts to ATP.Mg are released after adenylation to facilitate a 130 degree rotation of the Cys domain during thioester bond formation that is accompanied by remodelling of key structural elements including the helix that contains the E1 catalytic cysteine, the crossover and re-entry loops, and refolding of two helices that are required for adenylation. These changes displace side chains required for adenylation with side chains required for thioester bond formation. Mutational and biochemical analyses indicate these mechanisms are conserved in other E1s.
- Structural Biology, Sloan-Kettering Institute, New York, New York 10065, USA.
Organizational Affiliation: