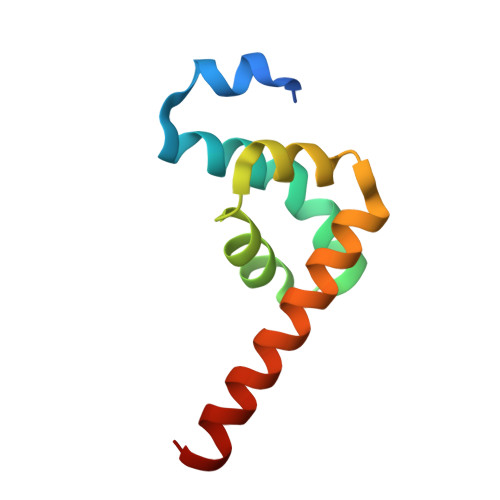
Molecular basis of eRF3 recognition by the MLLE domain of poly(A)-binding protein.
Kozlov, G., Gehring, K.(2010) PLoS One 5: e10169-e10169
- PubMed: 20418951
- DOI: https://doi.org/10.1371/journal.pone.0010169
- Primary Citation of Related Structures:
3KUI, 3KUJ - PubMed Abstract:
PABPC1 (cytosolic poly(A)-binding protein 1) is an RNA-binding protein that binds to the poly(A) tail of mRNAs to promote translation and mRNA turnover. In addition to RNA-binding domains, PABPC1 contains a unique protein-protein interaction domain, MLLE (also known as PABC) that binds regulatory proteins and translation factors that contain a conserved 12 amino acid peptide motif termed PAM2. Eukaryotic Release Factor 3 (eRF3/GSPT1) contains two overlapping PAM2 sequences, which are required for its activity. Here, we determined the crystal structures of the MLLE domain from PABPC1 in complex with the two PAM2 regions of eRF3. The structures reveal a mechanism of cooperativity between the two PAM2 sites that increases the binding affinity but prevents the binding of more than one molecule of eRF3 to PABPC1. Relative to previous structures, the high-resolution crystal structures force a re-evaluation of the PAM2 motif and improve our understanding of the molecular basis of MLLE peptide recognition.
- Department of Biochemistry, McGill University, Montréal, Québec, Canada.
Organizational Affiliation: