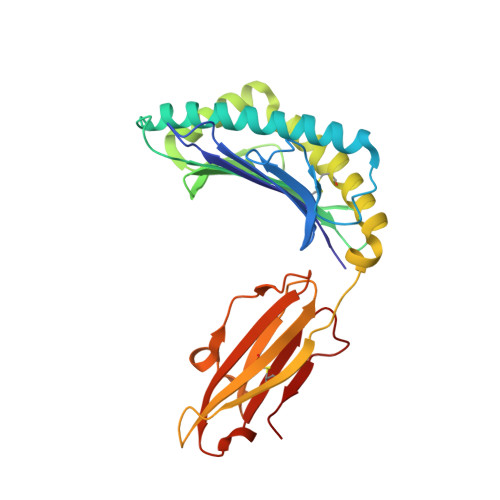
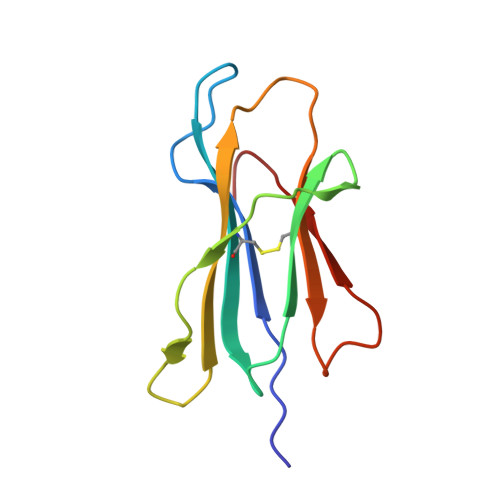
Ca2+ Release from the Endoplasmic Reticulum of NY-ESO-1-Specific T Cells Is Modulated by the Affinity of TCR and by the Use of the CD8 Coreceptor.
Chen, J.L., Morgan, A.J., Stewart-Jones, G., Shepherd, D., Bossi, G., Wooldridge, L., Hutchinson, S.L., Sewell, A.K., Griffiths, G.M., van der Merwe, P.A., Jones, E.Y., Galione, A., Cerundolo, V.(2010) J Immunol 184: 1829-1839
- PubMed: 20053942
- DOI: https://doi.org/10.4049/jimmunol.0902103
- Primary Citation of Related Structures:
3KLA - PubMed Abstract:
Although several cancer immunotherapy strategies are based on the use of analog peptides and on the modulation of the TCR affinity of adoptively transferred T cells, it remains unclear whether tumor-specific T cell activation by strong and weak TCR stimuli evoke different Ca(2+) signatures from the Ca(2+) intracellular stores and whether the amplitude of Ca(2+) release from the endoplasmic reticulum (ER) can be further modulated by coreceptor binding to peptide/MHC. In this study, we combined functional, structural, and kinetic measurements to correlate the intensity of Ca(2+) signals triggered by the stimulation of the 1G4 T cell clone specific to the tumor epitope NY-ESO-1(157-165). Two analogs of the NY-ESO-1(157-165) peptide, having similar affinity to HLA-A2 molecules, but a 6-fold difference in binding affinity for the 1G4 TCR, resulted in different Ca(2+) signals and T cell activation. 1G4 stimulation by the stronger stimulus emptied the ER of stored Ca(2+), even in the absence of CD8 binding, resulting in sustained Ca(2+) influx. In contrast, the weaker stimulus induced only partial emptying of stored Ca(2+), resulting in significantly diminished and oscillatory Ca(2+) signals, which were enhanced by CD8 binding. Our data define the range of TCR/peptide MHC affinities required to induce depletion of Ca(2+) from intracellular stores and provide insights into the ability of T cells to tailor the use of the CD8 coreceptor to enhance Ca(2+) release from the ER. This, in turn, modulates Ca(2+) influx from the extracellular environment, ultimately controlling T cell activation.
- Weatherall Institute of Molecular Medicine, OX3 9DS, Oxford.
Organizational Affiliation: