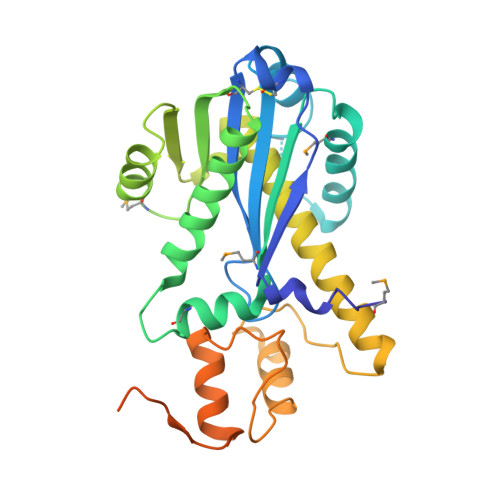
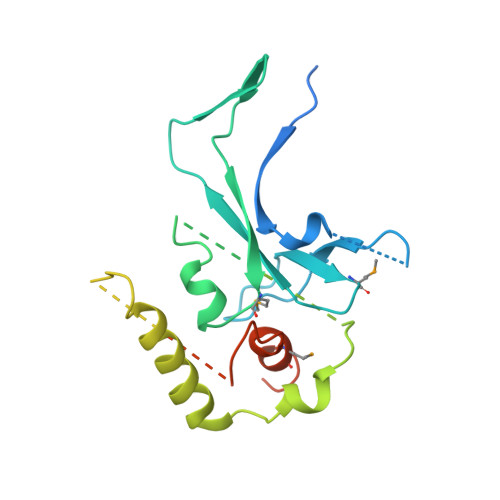
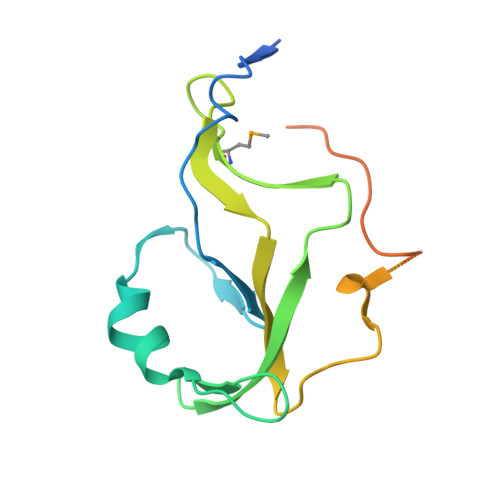
The structure of the mammalian RNase H2 complex provides insight into RNA:DNA hybrid processing to prevent immune dysfunction.
Shaban, N.M., Harvey, S., Perrino, F.W., Hollis, T.(2010) J Biological Chem 285: 3617-3624
- PubMed: 19923215
- DOI: https://doi.org/10.1074/jbc.M109.059048
- Primary Citation of Related Structures:
3KIO - PubMed Abstract:
The mammalian RNase H2 ribonuclease complex has a critical function in nucleic acid metabolism to prevent immune activation with likely roles in processing of RNA primers in Okazaki fragments during DNA replication, in removing ribonucleotides misinserted by DNA polymerases, and in eliminating RNA.DNA hybrids during cell death. Mammalian RNase H2 is a heterotrimeric complex of the RNase H2A, RNase H2B, and RNase H2C proteins that are all required for proper function and activity. Mutations in the human RNase H2 genes cause Aicardi-Goutières syndrome. We have determined the crystal structure of the three-protein mouse RNase H2 enzyme complex to better understand the molecular basis of RNase H2 dysfunction in human autoimmunity. The structure reveals the intimately interwoven architecture of RNase H2B and RNase H2C that interface with RNase H2A in a complex ideally suited for nucleic acid binding and hydrolysis coupled to protein-protein interaction motifs that could allow for efficient participation in multiple cellular functions. We have identified four conserved acidic residues in the active site that are necessary for activity and suggest a two-metal ion mechanism of catalysis for RNase H2. An Okazaki fragment has been modeled into the RNase H2 nucleic acid binding site providing insight into the recognition of RNA.DNA junctions by the RNase H2. Further structural and biochemical analyses show that some RNase H2 disease-causing mutations likely result in aberrant protein-protein interactions while the RNase H2A subunit-G37S mutation appears to distort the active site accounting for the demonstrated substrate specificity modification.
- From the Department of Biochemistry, Center for Structural Biology, Wake Forest University School of Medicine, Winston-Salem, North Carolina 27157.
Organizational Affiliation: