Crystal Structure of Glucocerebrosidase Containing the N370S Mutation, Implications for Chaperon Therapy
Wei, R.R., Boucher, S., Hughes, H., Guziewica, N., Pan, C.Q., Edmunds, T.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Glucosylceramidase | 497 | Homo sapiens | Mutation(s): 1 Gene Names: GBA, GC, GLUC EC: 3.2.1.45 (PDB Primary Data), 3.2.1.46 (UniProt), 2.4.1 (UniProt), 3.2.1 (UniProt) | 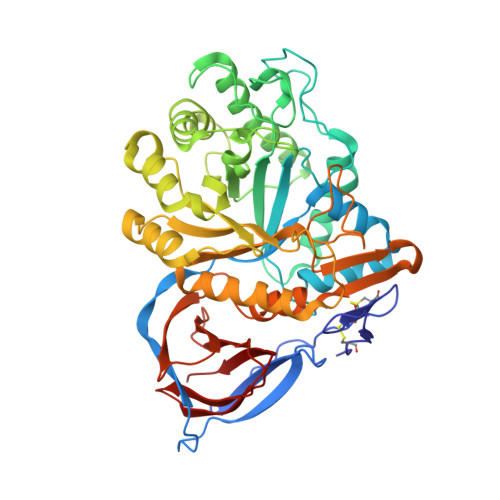 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P04062 (Homo sapiens) Explore P04062 Go to UniProtKB: P04062 | |||||
PHAROS: P04062 GTEx: ENSG00000177628 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P04062 | ||||
Glycosylation | |||||
| Glycosylation Sites: 2 | Go to GlyGen: P04062-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG Query on NAG | T [auth B] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| SO4 Query on SO4 | E [auth A] F [auth A] G [auth A] H [auth A] I [auth A] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| GOL Query on GOL | M [auth A], N [auth A], U [auth B] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 109.465 | α = 90 |
| b = 285.064 | β = 90 |
| c = 92.225 | γ = 90 |
| Software Name | Purpose |
|---|---|
| HKL-2000 | data collection |
| MOLREP | phasing |
| REFMAC | refinement |
| MOSFLM | data reduction |
| SCALA | data scaling |