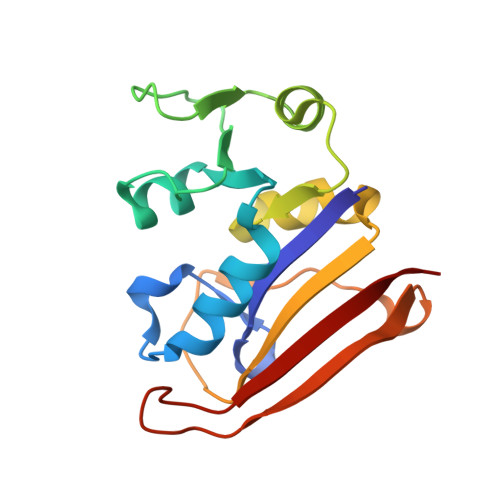
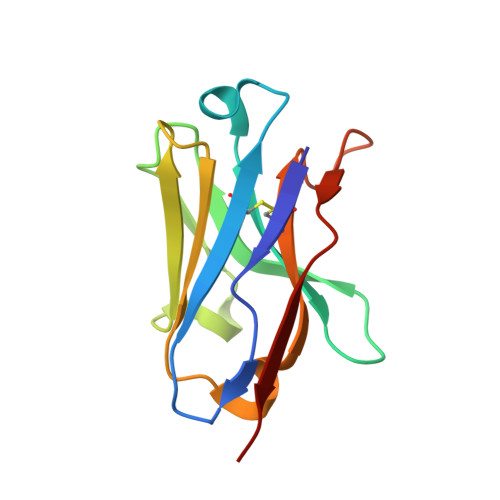
Constraining enzyme conformational change by an antibody leads to hyperbolic inhibition.
Oyen, D., Srinivasan, V., Steyaert, J., Barlow, J.N.(2011) J Mol Biology 407: 138-148
- PubMed: 21238460
- DOI: https://doi.org/10.1016/j.jmb.2011.01.017
- Primary Citation of Related Structures:
3K74 - PubMed Abstract:
Although it has been known for many years that antibodies display properties characteristic of allosteric effectors, the molecular mechanisms responsible for these effects remain poorly understood. Here, we describe a single-domain antibody fragment (nanobody) that modulates protein function by constraining conformational change in the enzyme dihydrofolate reductase (DHFR). Nanobody 216 (Nb216) behaves as a potent allosteric inhibitor of DHFR, giving rise to mixed hyperbolic inhibition kinetics. The crystal structure of Nb216 in complex with DHFR reveals that the nanobody binds adjacent to the active site. Half of the epitope consists of residues from the flexible Met20 loop. This loop, which ordinarily oscillates between occluded and closed conformations during catalysis, assumes the occluded conformation in the Nb216-bound state. Using stopped flow, we show that Nb216 inhibits DHFR by stabilising the occluded Met20 loop conformation. Surprisingly, kinetic data indicate that the Met20 loop retains sufficient conformational flexibility in the Nb216-bound state to allow slow substrate turnover to occur.
- Structural Biology Brussels, Vrije Universiteit Brussel, Pleinlaan 2, 1050 Brussels, Belgium.
Organizational Affiliation: