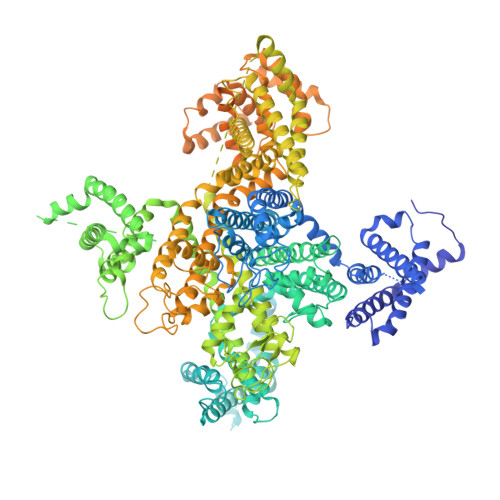
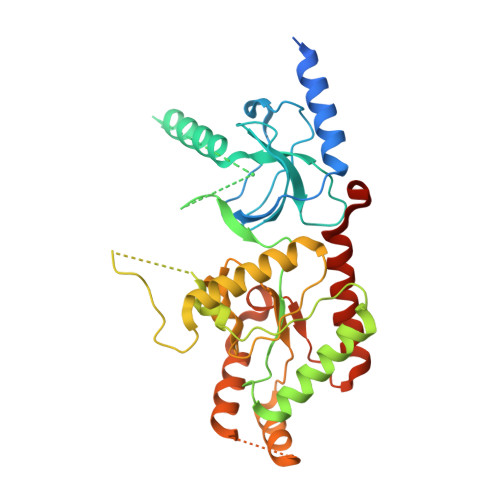
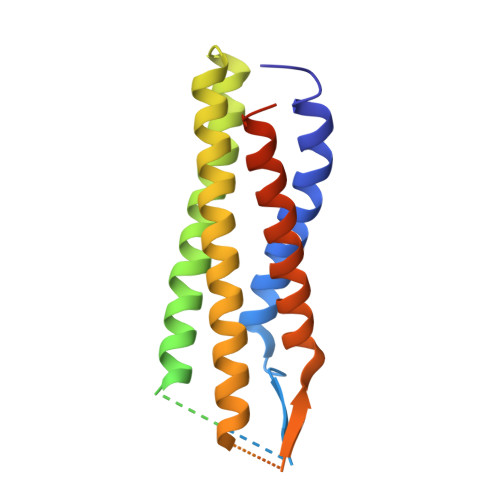
Structure of the voltage-gated calcium channel Cav1.1 complex
Wu, J.P., Yan, Z., Li, Z.Q., Yan, C.G., Lu, S., Dong, M.Q., Yan, N.(2015) Science 350: aad2395-aad2395
- PubMed: 26680202
- DOI: https://doi.org/10.1126/science.aad2395
- Primary Citation of Related Structures:
3JBR - PubMed Abstract:
The voltage-gated calcium channel Ca(v)1.1 is engaged in the excitation-contraction coupling of skeletal muscles. The Ca(v)1.1 complex consists of the pore-forming subunit α1 and auxiliary subunits α2δ, β, and γ. We report the structure of the rabbit Ca(v)1.1 complex determined by single-particle cryo-electron microscopy. The four homologous repeats of the α1 subunit are arranged clockwise in the extracellular view. The γ subunit, whose structure resembles claudins, interacts with the voltage-sensing domain of repeat IV (VSD(IV)), whereas the cytosolic β subunit is located adjacent to VSD(II) of α1. The α2 subunit interacts with the extracellular loops of repeats I to III through its VWA and Cache1 domains. The structure reveals the architecture of a prototypical eukaryotic Ca(v) channel and provides a framework for understanding the function and disease mechanisms of Ca(v) and Na(v) channels.
- State Key Laboratory of Membrane Biology, Tsinghua University, Beijing 100084, China. Tsinghua-Peking Joint Center for Life Sciences, Tsinghua University, Beijing 100084, China. Center for Structural Biology, School of Life Sciences and School of Medicine, Tsinghua University, Beijing 100084, China.
Organizational Affiliation: