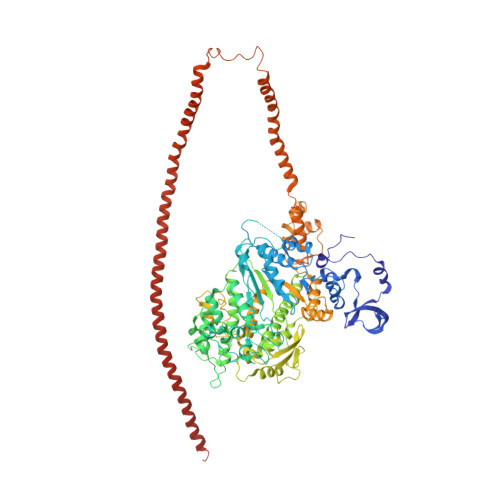
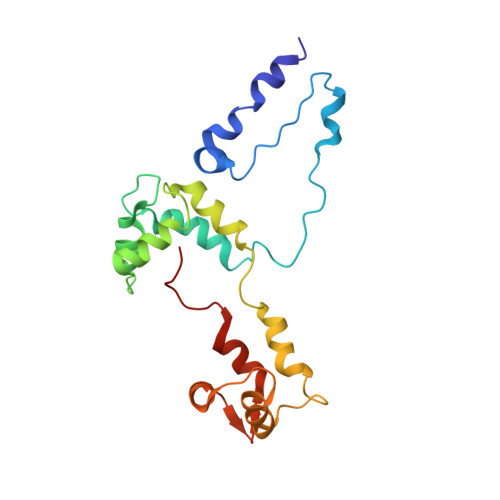
An invertebrate smooth muscle with striated muscle myosin filaments.
Sulbaran, G., Alamo, L., Pinto, A., Marquez, G., Mendez, F., Padron, R., Craig, R.(2015) Proc Natl Acad Sci U S A 112: E5660-E5668
- PubMed: 26443857
- DOI: https://doi.org/10.1073/pnas.1513439112
- Primary Citation of Related Structures:
3JAX - PubMed Abstract:
Muscle tissues are classically divided into two major types, depending on the presence or absence of striations. In striated muscles, the actin filaments are anchored at Z-lines and the myosin and actin filaments are in register, whereas in smooth muscles, the actin filaments are attached to dense bodies and the myosin and actin filaments are out of register. The structure of the filaments in smooth muscles is also different from that in striated muscles. Here we have studied the structure of myosin filaments from the smooth muscles of the human parasite Schistosoma mansoni. We find, surprisingly, that they are indistinguishable from those in an arthropod striated muscle. This structural similarity is supported by sequence comparison between the schistosome myosin II heavy chain and known striated muscle myosins. In contrast, the actin filaments of schistosomes are similar to those of smooth muscles, lacking troponin-dependent regulation. We conclude that schistosome muscles are hybrids, containing striated muscle-like myosin filaments and smooth muscle-like actin filaments in a smooth muscle architecture. This surprising finding has broad significance for understanding how muscles are built and how they evolved, and challenges the paradigm that smooth and striated muscles always have distinctly different components.
- Centro de Biología Estructural, Instituto Venezolano de Investigaciones Científicas (IVIC), Caracas 1020A, Venezuela; Department of Cell and Developmental Biology, University of Massachusetts Medical School, Worcester, MA 01655.
Organizational Affiliation: