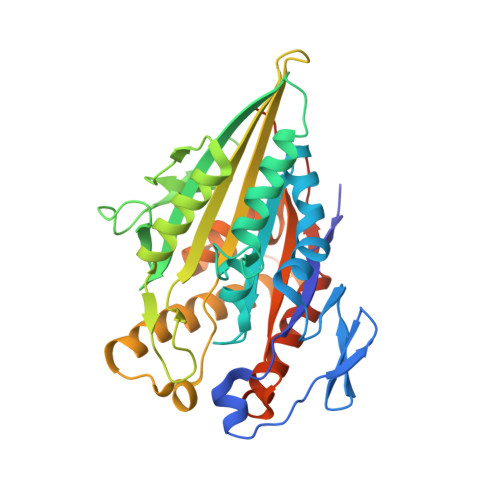
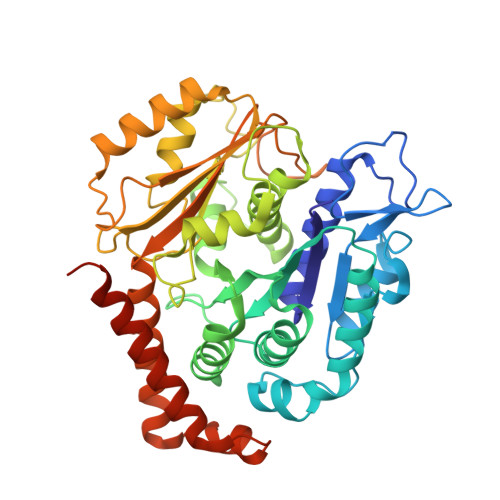
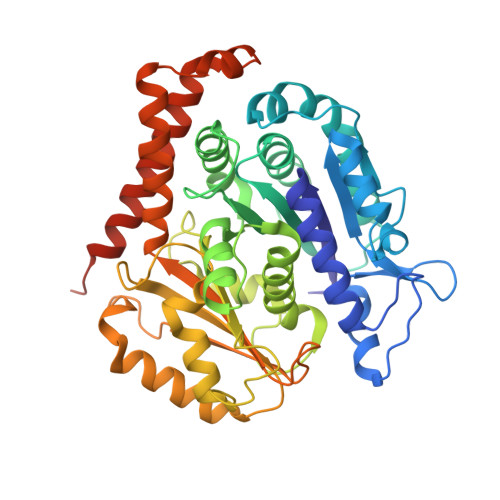
High-resolution structures of kinesin on microtubules provide a basis for nucleotide-gated force-generation.
Shang, Z., Zhou, K., Xu, C., Csencsits, R., Cochran, J.C., Sindelar, C.V.(2014) Elife 3: e04686-e04686
- PubMed: 25415053
- DOI: https://doi.org/10.7554/eLife.04686
- Primary Citation of Related Structures:
3J8X, 3J8Y - PubMed Abstract:
Microtubule-based transport by the kinesin motors, powered by ATP hydrolysis, is essential for a wide range of vital processes in eukaryotes. We obtained insight into this process by developing atomic models for no-nucleotide and ATP states of the monomeric kinesin motor domain on microtubules from cryo-EM reconstructions at 5-6 Å resolution. By comparing these models with existing X-ray structures of ADP-bound kinesin, we infer a mechanistic scheme in which microtubule attachment, mediated by a universally conserved 'linchpin' residue in kinesin (N255), triggers a clamshell opening of the nucleotide cleft and accompanying release of ADP. Binding of ATP re-closes the cleft in a manner that tightly couples to translocation of cargo, via kinesin's 'neck linker' element. These structural transitions are reminiscent of the analogous nucleotide-exchange steps in the myosin and F1-ATPase motors and inform how the two heads of a kinesin dimer 'gate' each other to promote coordinated stepping along microtubules.
- Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, United States.
Organizational Affiliation: