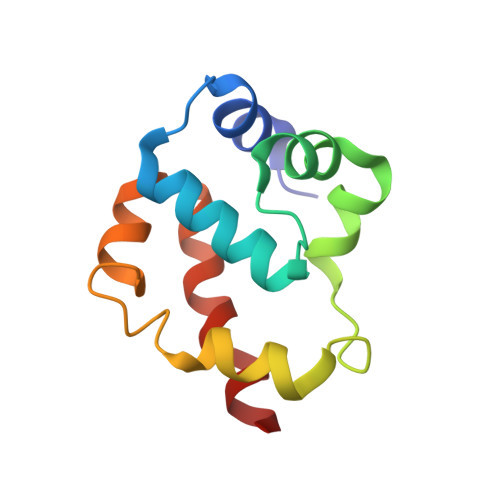
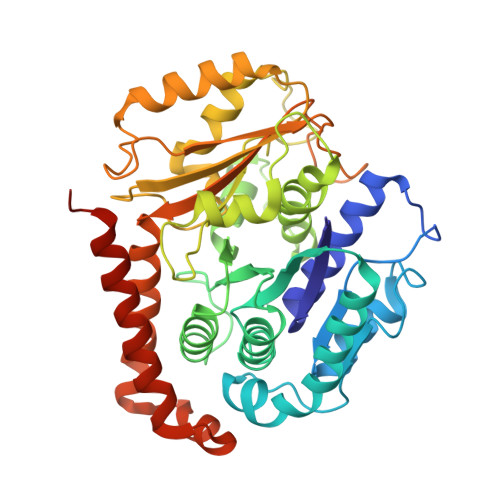
A flipped ion pair at the dynein-microtubule interface is critical for dynein motility and ATPase activation
Uchimura, S., Fujii, T., Takazaki, H., Ayukawa, R., Nishikawa, Y., Minoura, I., Hachikubo, Y., Kurisu, G., Sutoh, K., Kon, T., Namba, K., Muto, E.(2015) J Cell Biol 208: 211-222
- PubMed: 25583999
- DOI: https://doi.org/10.1083/jcb.201407039
- Primary Citation of Related Structures:
3J6P - PubMed Abstract:
Dynein is a motor protein that moves on microtubules (MTs) using the energy of adenosine triphosphate (ATP) hydrolysis. To understand its motility mechanism, it is crucial to know how the signal of MT binding is transmitted to the ATPase domain to enhance ATP hydrolysis. However, the molecular basis of signal transmission at the dynein-MT interface remains unclear. Scanning mutagenesis of tubulin identified two residues in α-tubulin, R403 and E416, that are critical for ATPase activation and directional movement of dynein. Electron cryomicroscopy and biochemical analyses revealed that these residues form salt bridges with the residues in the dynein MT-binding domain (MTBD) that work in concert to induce registry change in the stalk coiled coil and activate the ATPase. The R403-E3390 salt bridge functions as a switch for this mechanism because of its reversed charge relative to other residues at the interface. This study unveils the structural basis for coupling between MT binding and ATPase activation and implicates the MTBD in the control of directional movement.
- Laboratory for Molecular Biophysics, RIKEN Brain Science Institute, Wako, Saitama 351-0198, Japan.
Organizational Affiliation: