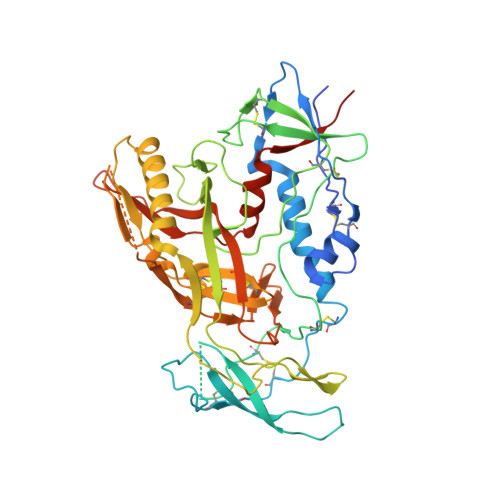
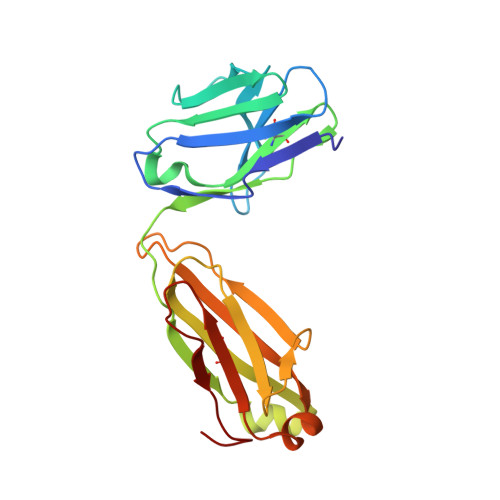
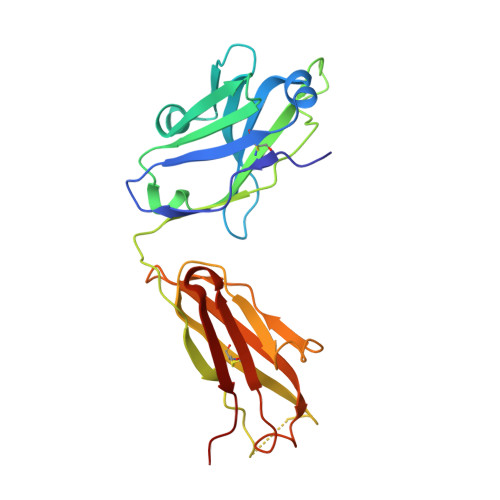
Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer.
Lyumkis, D., Julien, J.P., de Val, N., Cupo, A., Potter, C.S., Klasse, P.J., Burton, D.R., Sanders, R.W., Moore, J.P., Carragher, B., Wilson, I.A., Ward, A.B.(2013) Science 342: 1484-1490
- PubMed: 24179160
- DOI: https://doi.org/10.1126/science.1245627
- Primary Citation of Related Structures:
3J5M - PubMed Abstract:
The HIV-1 envelope glycoprotein (Env) trimer contains the receptor binding sites and membrane fusion machinery that introduce the viral genome into the host cell. As the only target for broadly neutralizing antibodies (bnAbs), Env is a focus for rational vaccine design. We present a cryo-electron microscopy reconstruction and structural model of a cleaved, soluble Env trimer (termed BG505 SOSIP.664 gp140) in complex with a CD4 binding site (CD4bs) bnAb, PGV04, at 5.8 angstrom resolution. The structure reveals the spatial arrangement of Env components, including the V1/V2, V3, HR1, and HR2 domains, as well as shielding glycans. The structure also provides insights into trimer assembly, gp120-gp41 interactions, and the CD4bs epitope cluster for bnAbs, which covers a more extensive area and defines a more complex site of vulnerability than previously described.
- National Resource for Automated Molecular Microscopy, The Scripps Research Institute, La Jolla, CA 92037, USA.
Organizational Affiliation: