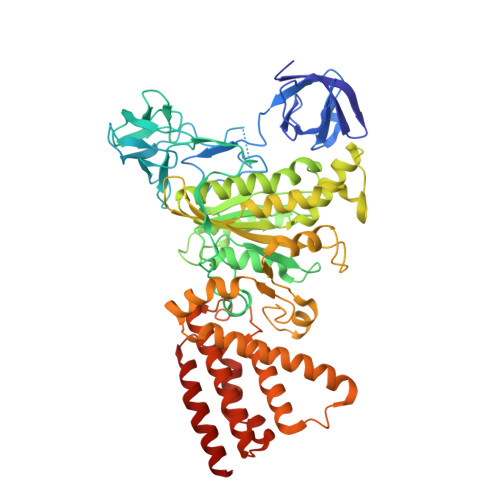
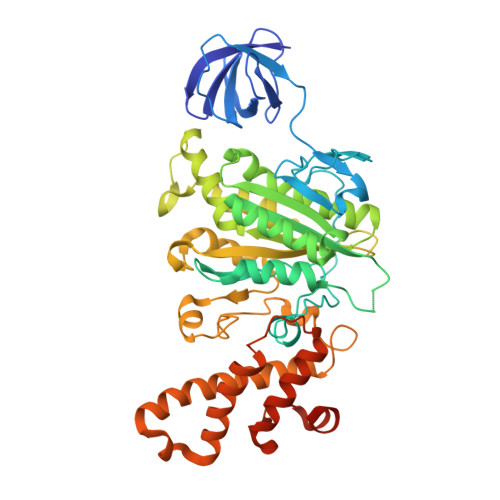
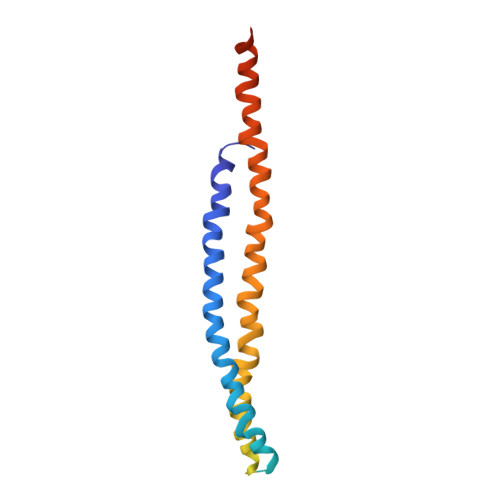
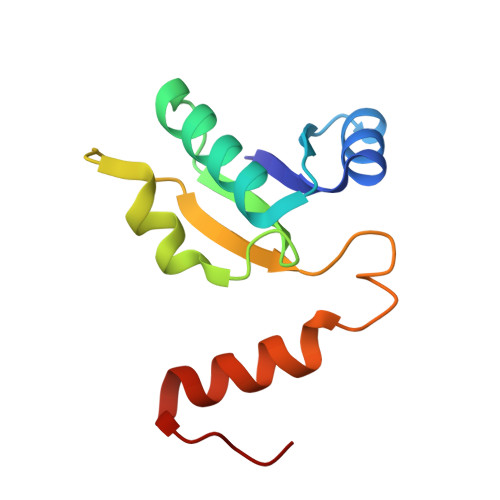
Subnanometre-resolution structure of the intact Thermus thermophilus H+-driven ATP synthase.
Lau, W.C., Rubinstein, J.L.(2012) Nature 481: 214-218
- PubMed: 22178924
- DOI: https://doi.org/10.1038/nature10699
- Primary Citation of Related Structures:
3J0J - PubMed Abstract:
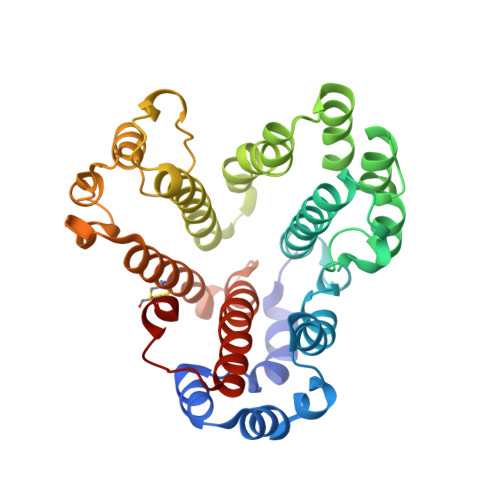
Ion-translocating rotary ATPases serve either as ATP synthases, using energy from a transmembrane ion motive force to create the cell's supply of ATP, or as transmembrane ion pumps that are powered by ATP hydrolysis. The members of this family of enzymes each contain two rotary motors: one that couples ion translocation to rotation and one that couples rotation to ATP synthesis or hydrolysis. During ATP synthesis, ion translocation through the membrane-bound region of the complex causes rotation of a central rotor that drives conformational changes and ATP synthesis in the catalytic region of the complex. There are no structural models available for the intact membrane region of any ion-translocating rotary ATPase. Here we present a 9.7 Å resolution map of the H(+)-driven ATP synthase from Thermus thermophilus obtained by electron cryomicroscopy of single particles in ice. The 600-kilodalton complex has an overall subunit composition of A(3)B(3)CDE(2)FG(2)IL(12). The membrane-bound motor consists of a ring of L subunits and the carboxy-terminal region of subunit I, which are equivalent to the c and a subunits of most other rotary ATPases, respectively. The map shows that the ring contains 12 L subunits and that the I subunit has eight transmembrane helices. The L(12) ring and I subunit have a surprisingly small contact area in the middle of the membrane, with helices from the I subunit making contacts with two different L subunits. The transmembrane helices of subunit I form bundles that could serve as half-channels across the membrane, with the first half-channel conducting protons from the periplasm to the L(12) ring and the second half-channel conducting protons from the L(12) ring to the cytoplasm. This structure therefore suggests the mechanism by which a transmembrane proton motive force is converted to rotation in rotary ATPases.
- Molecular Structure and Function Program, The Hospital for Sick Children Research Institute, 555 University Avenue, Toronto, Ontario M5G 1X8, Canada.
Organizational Affiliation: