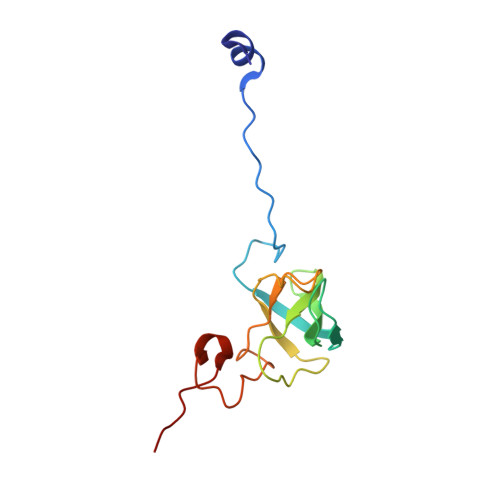
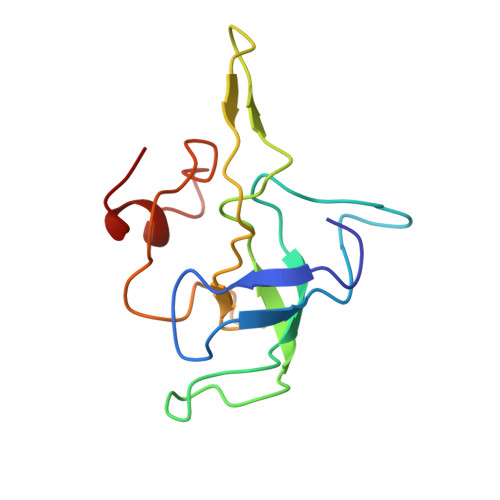
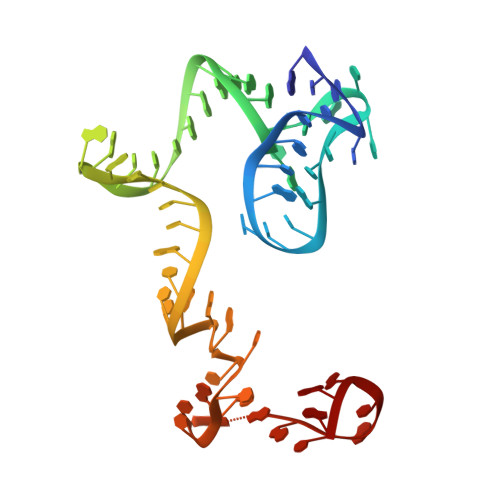
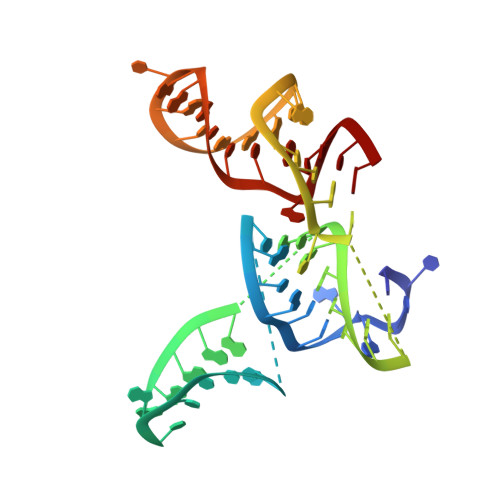Insertion domain within mammalian mitochondrial translation initiation factor 2 serves the role of eubacterial initiation factor 1.
Yassin, A.S., Haque, M.E., Datta, P.P., Elmore, K., Banavali, N.K., Spremulli, L.L., Agrawal, R.K.(2011) Proc Natl Acad Sci U S A 108: 3918-3923
- PubMed: 21368145
- DOI: https://doi.org/10.1073/pnas.1017425108
- Primary Citation of Related Structures:
3IZY, 3IZZ - PubMed Abstract:
Mitochondria have their own translational machineries for the synthesis of thirteen polypeptide chains that are components of the complexes that participate in the process of oxidative phosphorylation (or ATP generation). Translation initiation in mammalian mitochondria requires two initiation factors, IF2(mt) and IF3(mt), instead of the three that are present in eubacteria. The mammalian IF2(mt) possesses a unique 37 amino acid insertion domain, which is known to be important for the formation of the translation initiation complex. We have obtained a three-dimensional cryoelectron microscopic map of the mammalian IF2(mt) in complex with initiator fMet-tRNA(iMet) and the eubacterial ribosome. We find that the 37 amino acid insertion domain interacts with the same binding site on the ribosome that would be occupied by the eubacterial initiation factor IF1, which is absent in mitochondria. Our finding suggests that the insertion domain of IF2(mt) mimics the function of eubacterial IF1, by blocking the ribosomal aminoacyl-tRNA binding site (A site) at the initiation step.
- Wadsworth Center, New York State Department of Health, Empire State Plaza, Albany, NY 12201-0509, USA.
Organizational Affiliation: