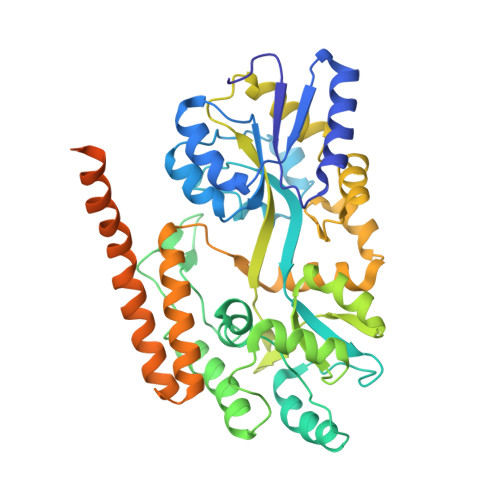
Secondary structure of Huntingtin amino-terminal region.
Kim, M.W., Chelliah, Y., Kim, S.W., Otwinowski, Z., Bezprozvanny, I.(2009) Structure 17: 1205-1212
- PubMed: 19748341
- DOI: https://doi.org/10.1016/j.str.2009.08.002
- Primary Citation of Related Structures:
3IO4, 3IO6, 3IOR, 3IOT, 3IOU, 3IOV, 3IOW - PubMed Abstract:
Huntington's disease is a genetic neurodegenerative disorder resulting from polyglutamine (polyQ) expansion (>36Q) within the first exon of Huntingtin (Htt) protein. We applied X-ray crystallography to determine the secondary structure of the first exon (EX1) of Htt17Q. The structure of Htt17Q-EX1 consists of an amino-terminal alpha helix, poly17Q region, and polyproline helix formed by the proline-rich region. The poly17Q region adopts multiple conformations in the structure, including alpha helix, random coil, and extended loop. The conformation of the poly17Q region is influenced by the conformation of neighboring protein regions, demonstrating the importance of the native protein context. We propose that the conformational flexibility of the polyQ region observed in our structure is a common characteristic of many amyloidogenic proteins. We further propose that the pathogenic polyQ expansion in the Htt protein increases the length of the random coil, which promotes aggregation and facilitates abnormal interactions with other proteins in cells.
- Department of Biochemistry, University of Texas Southwestern Medical Center at Dallas, Dallas, TX 75390, USA. MeeWhi.Kim@UTSouthwestern.edu
Organizational Affiliation: