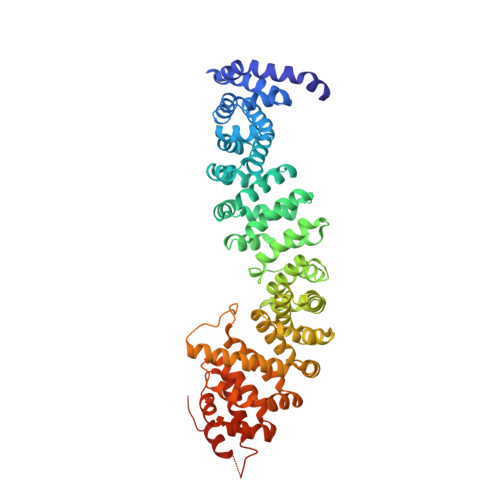
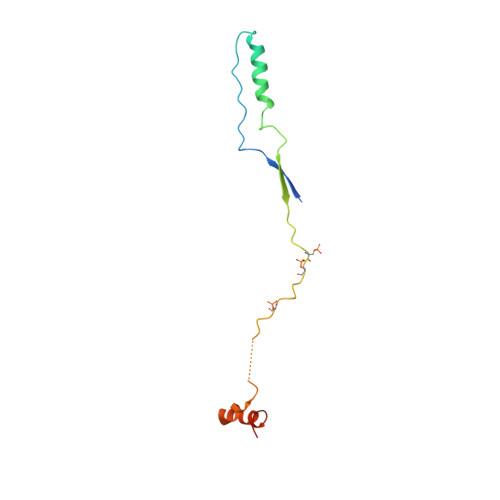
Interactions of plakoglobin and beta-catenin with desmosomal cadherins: basis of selective exclusion of alpha- and beta-catenin from desmosomes.
Choi, H.J., Gross, J.C., Pokutta, S., Weis, W.I.(2009) J Biological Chem 284: 31776-31788
- PubMed: 19759396
- DOI: https://doi.org/10.1074/jbc.M109.047928
- Primary Citation of Related Structures:
3IFQ - PubMed Abstract:
Plakoglobin and beta-catenin are homologous armadillo repeat proteins found in adherens junctions, where they interact with the cytoplasmic domain of classical cadherins and with alpha-catenin. Plakoglobin, but normally not beta-catenin, is also a structural constituent of desmosomes, where it binds to the cytoplasmic domains of the desmosomal cadherins, desmogleins and desmocollins. Here, we report structural, biophysical, and biochemical studies aimed at understanding the molecular basis of selective exclusion of beta-catenin and alpha-catenin from desmosomes. The crystal structure of the plakoglobin armadillo domain bound to phosphorylated E-cadherin shows virtually identical interactions to those observed between beta-catenin and E-cadherin. Trypsin sensitivity experiments indicate that the plakoglobin arm domain by itself is more flexible than that of beta-catenin. Binding of plakoglobin and beta-catenin to the intracellular regions of E-cadherin, desmoglein1, and desmocollin1 was measured by isothermal titration calorimetry. Plakoglobin and beta-catenin bind strongly and with similar thermodynamic parameters to E-cadherin. In contrast, beta-catenin binds to desmoglein-1 more weakly than does plakoglobin. beta-Catenin and plakoglobin bind with similar weak affinities to desmocollin-1. Full affinity binding of desmoglein-1 requires sequences C-terminal to the region homologous to the catenin-binding domain of classical cadherins. Although pulldown assays suggest that the presence of N- and C-terminal beta-catenin "tails" that flank the armadillo repeat region reduces the affinity for desmosomal cadherins, calorimetric measurements show no significant effects of the tails on binding to the cadherins. Using purified proteins, we show that desmosomal cadherins and alpha-catenin compete directly for binding to plakoglobin, consistent with the absence of alpha-catenin in desmosomes.
- Departments of Structural Biology and Molecular and Cellular Physiology, Stanford University School of Medicine, Stanford, California 94305-5126, USA.
Organizational Affiliation: