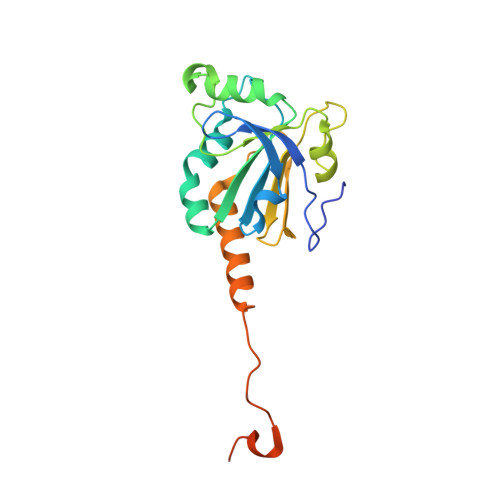
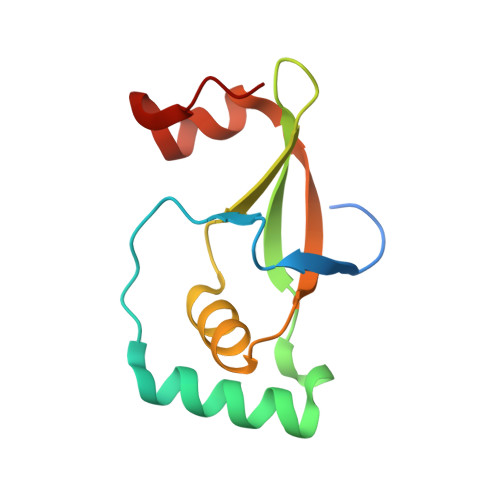
Protein Engineering of the Quaternary Sulfiredoxin-Peroxiredoxin Enzyme-Substrate Complex Reveals the Molecular Basis for Cysteine Sulfinic Acid Phosphorylation
Jonsson, T.J., Johnson, L.C., Lowther, W.T.(2009) J Biological Chem 284: 33305-33310
- PubMed: 19812042
- DOI: https://doi.org/10.1074/jbc.M109.036400
- Primary Citation of Related Structures:
3HY2 - PubMed Abstract:
Oxidative stress can damage the active site cysteine of the antioxidant enzyme peroxiredoxin (Prx) to the sulfinic acid form, Prx-SO(2)(-). This modification leads to inactivation. Sulfiredoxin (Srx) utilizes a unique ATP-Mg(2+)-dependent mechanism to repair the Prx molecule. Using selective protein engineering that involves disulfide bond formation and site-directed mutagenesis, a mimic of the enzyme.substrate complex has been trapped. Here, we present the 2.1 A crystal structure of human Srx in complex with PrxI, ATP, and Mg(2+). The Cys(52) sulfinic acid moiety was substituted by mutating this residue to Asp, leading to a replacement of the sulfur atom with a carbon atom. Because the Srx reaction cannot occur, the structural changes in the Prx active site that lead to the attack on ATP may be visualized. The local unfolding of the helix containing C52D resulted in the packing of Phe(50) in PrxI within a hydrophobic pocket of Srx. Importantly, this structural rearrangement positioned one of the oxygen atoms of Asp(52) within 4.3 A of the gamma-phosphate of ATP bound to Srx. These observations support a mechanism where phosphorylation of Prx-SO(2)(-) is the first chemical step.
- Center for Structural Biology and Department of Biochemistry, Wake Forest University School of Medicine, Winston-Salem, North Carolina 27157, USA.
Organizational Affiliation: