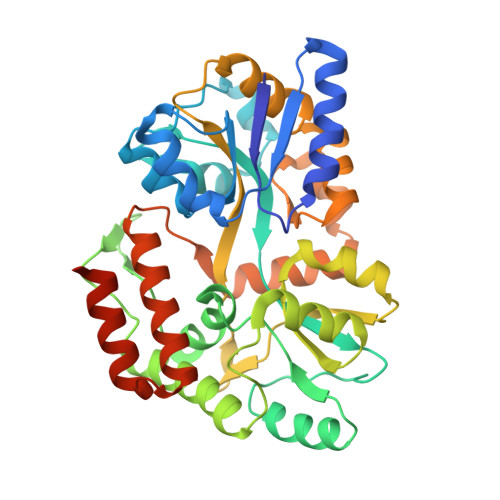
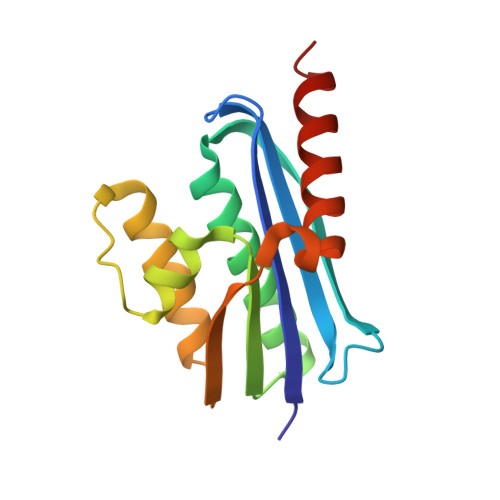
Structural and functional characterization of an RNase HI domain from the bifunctional protein Rv2228c from Mycobacterium tuberculosis.
Watkins, H.A., Baker, E.N.(2010) J Bacteriol 192: 2878-2886
- PubMed: 20363939
- DOI: https://doi.org/10.1128/JB.01615-09
- Primary Citation of Related Structures:
3HST - PubMed Abstract:
The open reading frame Rv2228c from Mycobacterium tuberculosis is predicted to encode a protein composed of two domains, each with individual functions, annotated through sequence similarity searches. The N-terminal domain is homologous with prokaryotic and eukaryotic RNase H domains and the C-terminal domain with alpha-ribazole phosphatase (CobC). The N-terminal domain of Rv2228c (Rv2228c/N) and the full-length protein were expressed as fusions with maltose binding protein (MBP). Rv2228c/N was shown to have RNase H activity with a hybrid RNA/DNA substrate as well as double-stranded RNase activity. The full-length protein was shown to have additional CobC activity. The crystal structure of the MBP-Rv2228c/N fusion protein was solved by molecular replacement and refined at 2.25-A resolution (R = 0.182; R(free) = 0.238). The protein is monomeric in solution but associates in the crystal to form a dimer. The Rv2228c/N domain has the classic RNase H fold and catalytic machinery but lacks several surface features that play important roles in the cleavage of RNA/DNA hybrids by other RNases H. The absence of either the basic protrusion of some RNases H or the hybrid binding domain of others appears to be compensated by the C-terminal CobC domain in full-length Rv2228c. The double-stranded-RNase activity of Rv2228c/N contrasts with classical RNases H and is attributed to the absence in Rv2228c/N of a key phosphate binding pocket.
- Maurice Wilkins Centre for Molecular Biodiscovery and School of Biological Sciences, 3A Symonds Street, Private Bag 92019, Auckland, New Zealand.
Organizational Affiliation: