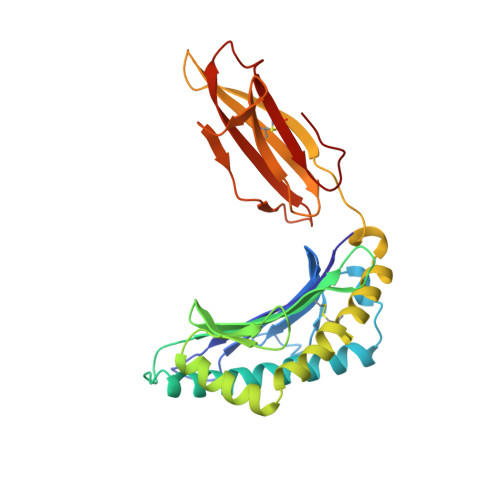
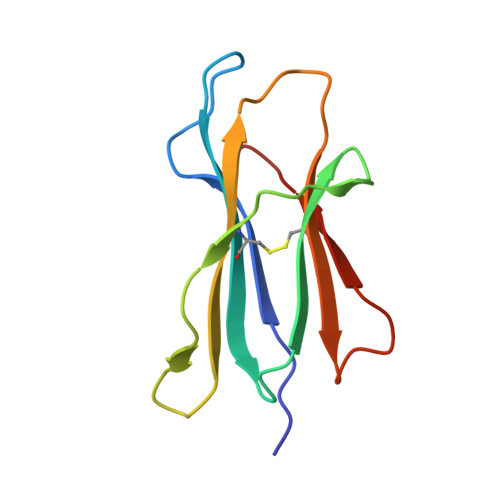
Refined structure of the human histocompatibility antigen HLA-A2 at 2.6 A resolution.
Saper, M.A., Bjorkman, P.J., Wiley, D.C.(1991) J Mol Biology 219: 277-319
- PubMed: 2038058
- DOI: https://doi.org/10.1016/0022-2836(91)90567-p
- Primary Citation of Related Structures:
3HLA - PubMed Abstract:
The three-dimensional structure of the human histocompatibility antigen HLA-A2 was determined at 3.5 A resolution by a combination of isomorphous replacement and iterative real-space averaging of two crystal forms. The monoclinic crystal form has now been refined by least-squares methods to an R-factor of 0.169 for data from 6 to 2.6 A resolution. A superposition of the structurally similar domains found in the heterodimer, alpha 1 onto alpha 2 and alpha 3 onto beta 2m, as well as the latter pair onto the ancestrally related immunoglobulin constant domain, reveals that differences are mainly in the turn regions. Structural features of the alpha 1 and alpha 2 domains, such as conserved salt-bridges that contribute to stability, specific loops that form contacts with other domains, and the antigen-binding groove formed from two adjacent helical regions on top of an eight-stranded beta-sheet, are analyzed. The interfaces between the domains, especially those between beta 2m and the HLA heavy chain presumably involved in beta 2m exchange and heterodimer assembly, are described in detail. A detailed examination of the binding groove confirms that the solvent-accessible amino acid side-chains that are most polymorphic in mouse and human alleles fill up the central and widest portion of the binding groove, while conserved side-chains are clustered at the narrower ends of the groove. Six pockets or sub-sites in the antigen-binding groove, of diverse shape and composition, appear suited for binding side-chains from antigenic peptides. Three pockets contain predominantly non-polar atoms; but others, especially those at the extreme ends of the groove, have clusters of polar atoms in close proximity to the "extra" electron density in the binding site. A possible role for beta 2m in stabilizing permissible peptide complexes during folding and assembly is presented.
- Department of Biochemistry and Molecular Biology, Howard Hughes Medical Institute, Harvard University, Cambridge, MA 02138.
Organizational Affiliation: