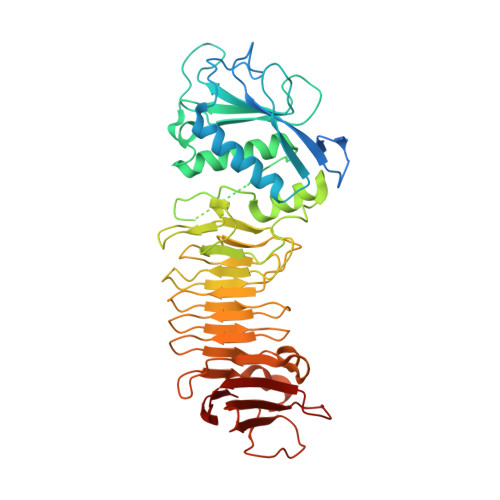
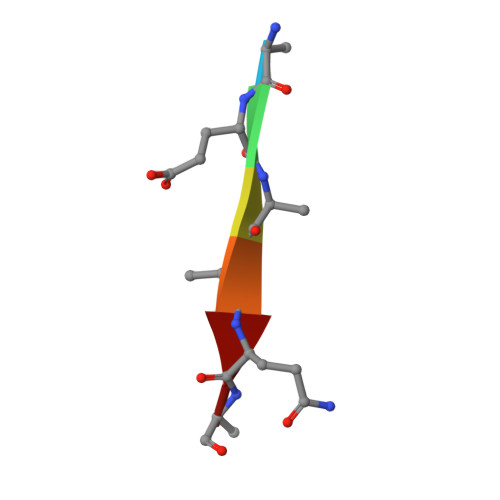
Metzincin's canonical methionine is responsible for the structural integrity of the zinc-binding site
Oberholzer, A.E., Bumann, M., Hege, T., Russo, S., Baumann, U.(2009) Biol Chem 390: 875-881
- PubMed: 19558324
- DOI: https://doi.org/10.1515/BC.2009.100
- Primary Citation of Related Structures:
3HB2, 3HBU, 3HBV, 3HDA - PubMed Abstract:
The metzincins constitute a subclan of metalloproteases possessing a HEXXHXXGXXH/D zinc-binding consensus sequence where the three histidines are zinc ligands and the glutamic acid is the catalytic base. A completely conserved methionine is located downstream of this motif. Families of the metzincin clan comprise, besides others, astacins, adamalysins proteases, matrix metallo-proteases, and serralysins. The latter are extracellular 50 kDa proteases secreted by Gram-negative bacteria via a type I secretion system. While there is a large body of structural and biochemical information available, the function of the conserved methionine has not been convincingly clarified yet. Here, we present the crystal structures of a number of mutants of the serralysin member protease C with the conserved methionine being replaced by Ile, Ala, and His. Together with our former report on the leucine and cysteine mutants, we demonstrate here that replacement of the methionine side chain results in an increasing distortion of the zinc-binding geometry, especially pronounced in the chi(2) angles of the first and third histidine of the consensus sequence. This is correlated with an increasing loss of proteolytic activity and a sharp increase of flexibility of large segments of the polypeptide chain.
- Department of Chemistry and Biochemistry, University of Bern, Bern, Switzerland.
Organizational Affiliation: