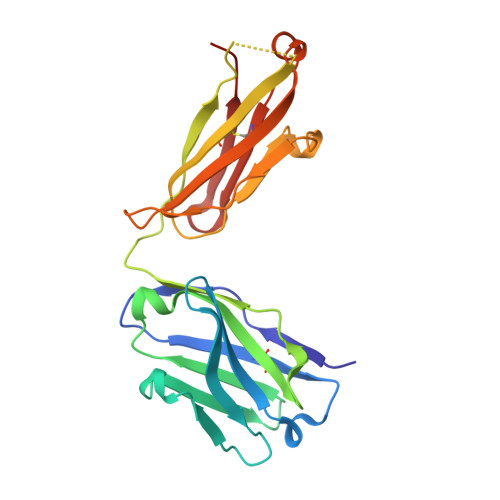
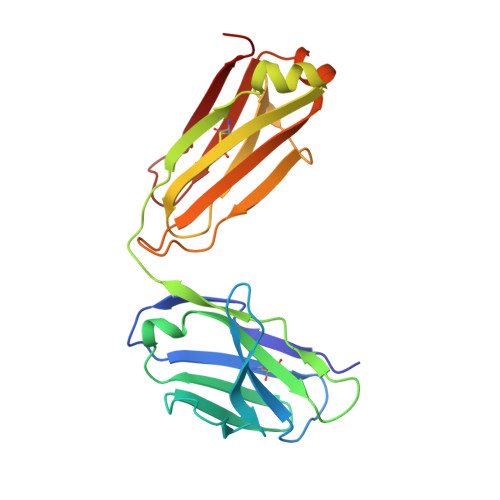
Structural understanding of stabilization patterns in engineered bispecific Ig-like antibody molecules
Jordan, J.L., Arndt, J.W., Hanf, K., Li, G., Hall, J., Demarest, S., Huang, F., Wu, X., Miller, B., Glaser, S., Fernandez, E.J., Wang, D., Lugovskoy, A.(2009) Proteins 77: 832-841
- PubMed: 19626705
- DOI: https://doi.org/10.1002/prot.22502
- Primary Citation of Related Structures:
3HC0, 3HC3, 3HC4 - PubMed Abstract:
Bispecific immunoglobulin-like antibodies capable of engaging multiple antigens represent a promising new class of therapeutic agents. Engineering of these molecules requires optimization of the molecular properties of one of the domain components. Here, we present a detailed crystallographic and computational characterization of the stabilization patterns in the lymphotoxin-beta receptor (LTbetaR) binding Fv domain of an anti-LTbetaR/anti-TNF-related apoptosis inducing ligand receptor-2 (TRAIL-R2) bispecific immunoglobulin-like antibody. We further describe a new hierarchical structure-guided approach toward engineering of antibody-like molecules to enhance their thermal and chemical stability.
- Biogen Idec, Inc., 12 Cambridge Center, Cambridge, Massachusetts 02142, USA.
Organizational Affiliation: