Structural Insights into the Catalytic Mechanism of Arabidopsis thaliana Agmatine Deiminase
Burgie, E.S., Bingman, C.A., Phillips Jr., G.N.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Agmatine deiminase | A [auth X] | 383 | Arabidopsis thaliana | Mutation(s): 0 Gene Names: AIH, At5g08170, EMB1873, T22D6.110 EC: 3.5.3.12 | 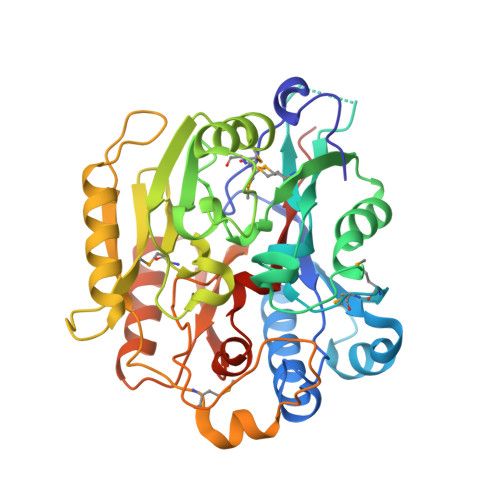 |
UniProt | |||||
Find proteins for Q8GWW7 (Arabidopsis thaliana) Explore Q8GWW7 Go to UniProtKB: Q8GWW7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8GWW7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 6 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| 1PE Query on 1PE | I [auth X] | PENTAETHYLENE GLYCOL C10 H22 O6 JLFNLZLINWHATN-UHFFFAOYSA-N |  | ||
| 211 Query on 211 | H [auth X] | 2,2',2''-NITRILOTRIETHANOL C6 H15 N O3 GSEJCLTVZPLZKY-UHFFFAOYSA-N |  | ||
| K Query on K | G [auth X] | POTASSIUM ION K NPYPAHLBTDXSSS-UHFFFAOYSA-N |  | ||
| CL Query on CL | B [auth X], C [auth X] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| MG Query on MG | D [auth X] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| NA Query on NA | E [auth X], F [auth X] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| MSE Query on MSE | A [auth X] | L-PEPTIDE LINKING | C5 H11 N O2 Se |  | MET |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 123.715 | α = 90 |
| b = 69.932 | β = 98.4 |
| c = 50.917 | γ = 90 |
| Software Name | Purpose |
|---|---|
| DENZO | data reduction |
| SCALEPACK | data scaling |
| RESOLVE | phasing |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| HKL-2000 | data collection |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| AutoSol | phasing |