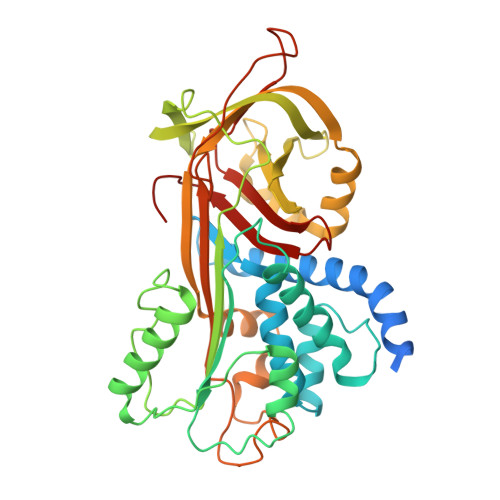
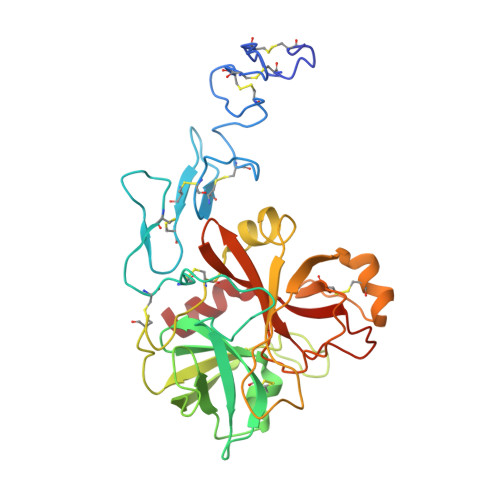
Basis for the specificity and activation of the serpin protein Z-dependent proteinase inhibitor (ZPI) as an inhibitor of membrane-associated factor Xa.
Huang, X., Dementiev, A., Olson, S.T., Gettins, P.G.(2010) J Biological Chem 285: 20399-20409
- PubMed: 20427285
- DOI: https://doi.org/10.1074/jbc.M110.112748
- Primary Citation of Related Structures:
3H5C - PubMed Abstract:
The serpin ZPI is a protein Z (PZ)-dependent specific inhibitor of membrane-associated factor Xa (fXa) despite having an unfavorable P1 Tyr. PZ accelerates the inhibition reaction approximately 2000-fold in the presence of phospholipid and Ca(2+). To elucidate the role of PZ, we determined the x-ray structure of Gla-domainless PZ (PZ(DeltaGD)) complexed with protein Z-dependent proteinase inhibitor (ZPI). The PZ pseudocatalytic domain bound ZPI at a novel site through ionic and polar interactions. Mutation of four ZPI contact residues eliminated PZ binding and membrane-dependent PZ acceleration of fXa inhibition. Modeling of the ternary Michaelis complex implicated ZPI residues Glu-313 and Glu-383 in fXa binding. Mutagenesis established that only Glu-313 is important, contributing approximately 5-10-fold to rate acceleration of fXa and fXIa inhibition. Limited conformational change in ZPI resulted from PZ binding, which contributed only approximately 2-fold to rate enhancement. Instead, template bridging from membrane association, together with previously demonstrated interaction of the fXa and ZPI Gla domains, resulted in an additional approximately 1000-fold rate enhancement. To understand why ZPI has P1 tyrosine, we examined a P1 Arg variant. This reacted at a diffusion-limited rate with fXa, even without PZ, and predominantly as substrate, reflecting both rapid acylation and deacylation. P1 tyrosine thus ensures that reaction with fXa or most other arginine-specific proteinases is insignificant unless PZ binds and localizes ZPI and fXa on the membrane, where the combined effects of Gla-Gla interaction, template bridging, and interaction of fXa with Glu-313 overcome the unfavorability of P1 Tyr and ensure a high rate of reaction as an inhibitor.
- Center for Molecular Biology of Oral Diseases, University of Illinois at Chicago, Chicago, Illinois 60607, USA.
Organizational Affiliation: