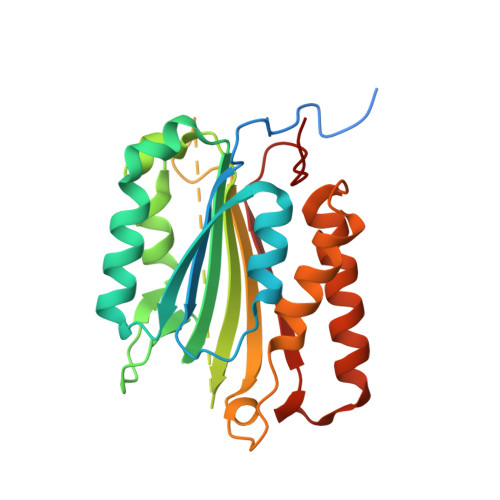
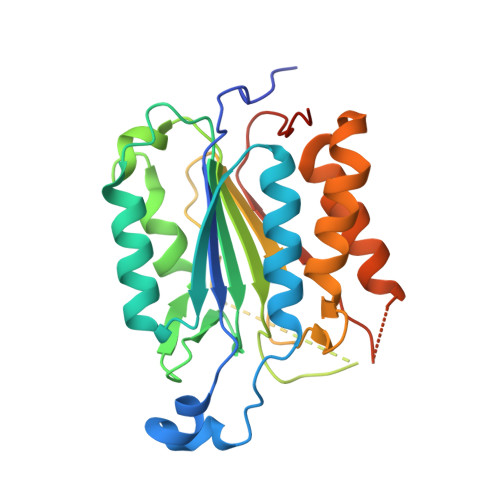
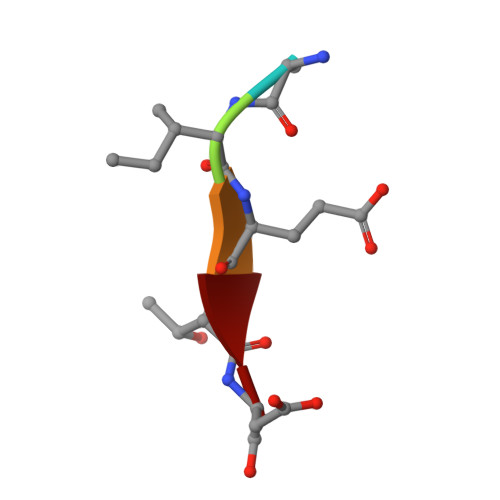
Mechanism of procaspase-8 activation by c-FLIPL.
Yu, J.W., Jeffrey, P.D., Shi, Y.(2009) Proc Natl Acad Sci U S A 106: 8169-8174
- PubMed: 19416807
- DOI: https://doi.org/10.1073/pnas.0812453106
- Primary Citation of Related Structures:
3H11, 3H13 - PubMed Abstract:
Cellular FLICE-inhibitory protein (c-FLIP(L)) is a key regulator of the extrinsic cell death pathway. Although widely regarded as an inhibitor of initiator caspase activation and cell death, c-FLIP(L) is also capable of enhancing procaspase-8 activation through heterodimerization of their respective protease domains. However, the underlying mechanism of this activation process remains enigmatic. Here, we demonstrate that cleavage of the intersubunit linker of c-FLIP(L) by procaspase-8 potentiates the activation process by enhancing heterodimerization between the two proteins and vastly improving the proteolytic activity of unprocessed caspase-(C)8. The crystal structures of the protease-like domain of c-FLIP(L) alone and in complex with zymogen C8 identify the unique determinants that favor heterodimerization over procaspase-8 homodimerization, and induce the latent active site of zymogen C8 into a productive conformation. Together, these findings provide molecular insights into a key aspect of c-FLIP(L) function that modulates procaspase-8 activation to elicit diverse responses in different cellular contexts.
- Department of Molecular Biology, Lewis Thomas Laboratory, Princeton University, Princeton, NJ 08544, USA. jwyu@princeton.edu
Organizational Affiliation: