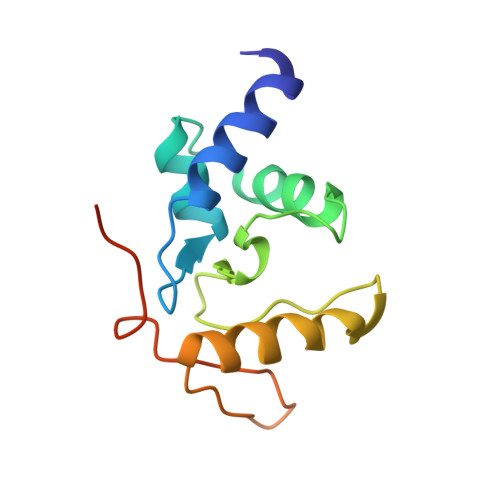
Flipping of alkylated DNA damage bridges base and nucleotide excision repair.
Tubbs, J.L., Latypov, V., Kanugula, S., Butt, A., Melikishvili, M., Kraehenbuehl, R., Fleck, O., Marriott, A., Watson, A.J., Verbeek, B., McGown, G., Thorncroft, M., Santibanez-Koref, M.F., Millington, C., Arvai, A.S., Kroeger, M.D., Peterson, L.A., Williams, D.M., Fried, M.G., Margison, G.P., Pegg, A.E., Tainer, J.A.(2009) Nature 459: 808-813
- PubMed: 19516334
- DOI: https://doi.org/10.1038/nature08076
- Primary Citation of Related Structures:
3GVA, 3GX4, 3GYH - PubMed Abstract:
Alkyltransferase-like proteins (ATLs) share functional motifs with the cancer chemotherapy target O(6)-alkylguanine-DNA alkyltransferase (AGT) and paradoxically protect cells from the biological effects of DNA alkylation damage, despite lacking the reactive cysteine and alkyltransferase activity of AGT. Here we determine Schizosaccharomyces pombe ATL structures without and with damaged DNA containing the endogenous lesion O(6)-methylguanine or cigarette-smoke-derived O(6)-4-(3-pyridyl)-4-oxobutylguanine. These results reveal non-enzymatic DNA nucleotide flipping plus increased DNA distortion and binding pocket size compared to AGT. Our analysis of lesion-binding site conservation identifies new ATLs in sea anemone and ancestral archaea, indicating that ATL interactions are ancestral to present-day repair pathways in all domains of life. Genetic connections to mammalian XPG (also known as ERCC5) and ERCC1 in S. pombe homologues Rad13 and Swi10 and biochemical interactions with Escherichia coli UvrA and UvrC combined with structural results reveal that ATLs sculpt alkylated DNA to create a genetic and structural intersection of base damage processing with nucleotide excision repair.
- Skaggs Institute for Chemical Biology and Department of Molecular Biology, The Scripps Research Institute, La Jolla, California 92037, USA.
Organizational Affiliation: