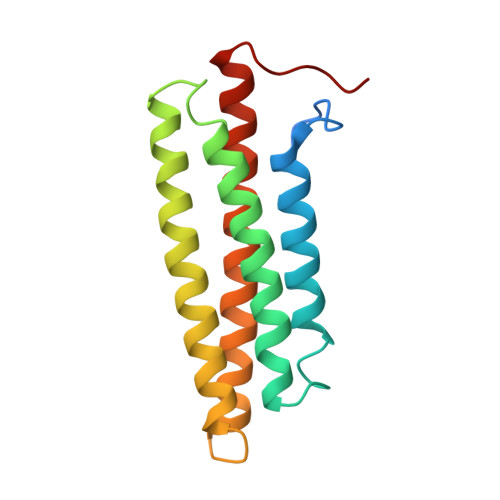
Crystal structures of free and ligand-bound focal adhesion targeting domain of Pyk2
Lulo, J., Yuzawa, S., Schlessinger, J.(2009) Biochem Biophys Res Commun 383: 347-352
- PubMed: 19358827
- DOI: https://doi.org/10.1016/j.bbrc.2009.04.011
- Primary Citation of Related Structures:
3GM1, 3GM2, 3GM3 - PubMed Abstract:
Focal adhesion targeting (FAT) domains target the non-receptor tyrosine kinases FAK and Pyk2 to cellular focal adhesion areas, where the signaling molecule paxillin is also located. Here, we report the crystal structures of the Pyk2 FAT domain alone or in complex with paxillin LD4 peptides. The overall structure of Pyk2-FAT is an antiparallel four-helix bundle with an up-down, up-down, right-handed topology. In the LD4-bound FAT complex, two paxillin LD4 peptides interact with two opposite sides of Pyk2-FAT, at the surfaces of the alpha1alpha4 and alpha2alpha3 helices of each FAT molecule. We also demonstrate that, while paxillin is phosphorylated by Pyk2, complex formation between Pyk2 and paxillin does not depend on Pyk2 tyrosine kinase activity. These experiments reveal the structural basis underlying the selectivity of paxillin LD4 binding to the Pyk2 FAT domain and provide insights about the molecular details which influence the different behavior of these two closely-related kinases.
- Department of Pharmacology, Yale University School of Medicine, New Haven, CT 06520, USA.
Organizational Affiliation: