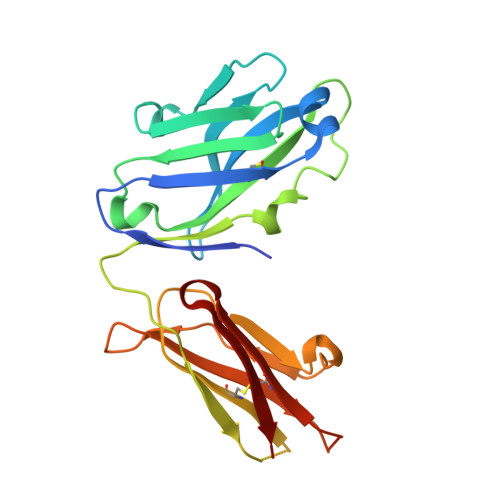
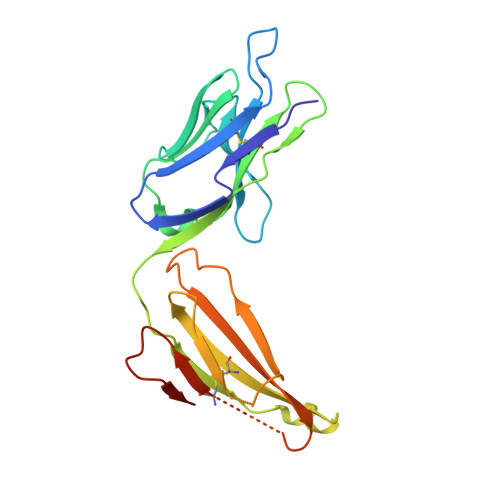
Nimotuzumab, an antitumor antibody that targets the epidermal growth factor receptor, blocks ligand binding while permitting the active receptor conformation
Talavera, A., Friemann, R., Gomez-Puerta, S., Martinez-Fleites, C., Garrido, G., Rabasa, A., Lopez-Requena, A., Pupo, A., Johansen, R.F., Sanchez, O., Krengel, U., Moreno, E.(2009) Cancer Res 69: 5851-5859
- PubMed: 19584289
- DOI: https://doi.org/10.1158/0008-5472.CAN-08-4518
- Primary Citation of Related Structures:
3GKW - PubMed Abstract:
Overexpression of the epidermal growth factor (EGF) receptor (EGFR) in cancer cells correlates with tumor malignancy and poor prognosis for cancer patients. For this reason, the EGFR has become one of the main targets of anticancer therapies. Structural data obtained in the last few years have revealed the molecular mechanism for ligand-induced EGFR dimerization and subsequent signal transduction, and also how this signal is blocked by either monoclonal antibodies or small molecules. Nimotuzumab (also known as h-R3) is a humanized antibody that targets the EGFR and has been successful in the clinics. In this work, we report the crystal structure of the Fab fragment of Nimotuzumab, revealing some unique structural features in the heavy variable domain. Furthermore, competition assays show that Nimotuzumab binds to domain III of the extracellular region of the EGFR, within an area that overlaps with both the surface patch recognized by Cetuximab (another anti-EGFR antibody) and the binding site for EGF. A computer model of the Nimotuzumab-EGFR complex, constructed by docking and molecular dynamics simulations and supported by mutagenesis studies, unveils a novel mechanism of action, with Nimotuzumab blocking EGF binding while still allowing the receptor to adopt its active conformation, hence warranting a basal level of signaling.
- Center of Molecular Immunology, Havana, Cuba.
Organizational Affiliation: