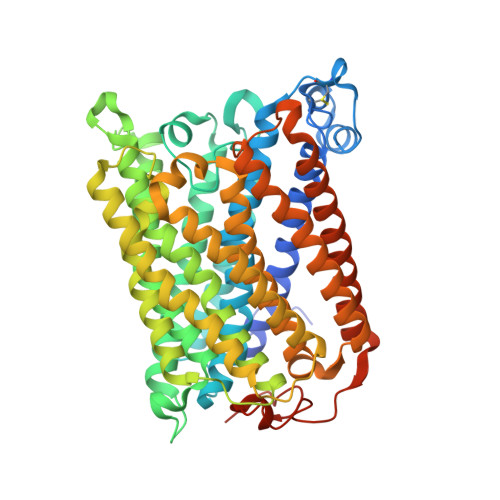
Redox dependent conformational changes in cytochrome c oxidase suggest a gating mechanism for proton uptake.
Qin, L., Liu, J., Mills, D., Proshlyakov, D.A., Hiser, C., Ferguson-Miller, S.(2009) Biochemistry 48: 5121-5130
- PubMed: 19397279
- DOI: https://doi.org/10.1021/bi9001387
- Primary Citation of Related Structures:
3FYE, 3FYI - PubMed Abstract:
A role for conformational change in the coupling mechanism of cytochrome c oxidase is the subject of controversy. Relatively small conformational changes have been reported in comparisons of reduced and oxidized crystal structures of bovine oxidase but none in bacterial oxidases. Comparing the X-ray crystal structures of the reduced (at 2.15 A resolution) and oxidized forms of cytochrome c oxidase from Rhodobacter sphaeroides, we observe a displacement of heme a(3) involving both the porphyrin ring and the hydroxyl farnesyl tail, accompanied by protein movements in nearby regions, including the mid part of helix VIII of subunit I which harbors key residues of the K proton uptake path, K362 and T359. The conformational changes in the reduced form are reversible upon reoxidation. They result in an opening of the top of the K pathway and more ordered waters being resolved in that region, suggesting an access path for protons into the active site. In all high-resolution structures of oxidized R. sphaeroides cytochrome c oxidase, a water molecule is observed in the hydrophobic region above the top of the D path, strategically positioned to facilitate the connection of residue E286 of subunit I to the active site or to the proton pumping exit path. In the reduced and reduced plus cyanide structures, this water molecule disappears, implying disruption of proton conduction from the D path under conditions when the K path is open, thus providing a mechanism for alternating access to the active site.
- Biochemistry and Molecular Biology Department, Michigan State University, East Lansing, Michigan 48824, USA.
Organizational Affiliation: