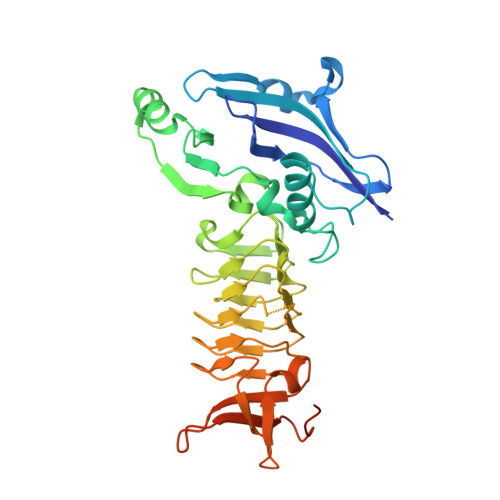
The three-dimensional Structure of a mycobacterial DapD provides insights into DapD diversity and reveals unexpected particulars about the enzymatic mechanism.
Schuldt, L., Weyand, S., Kefala, G., Weiss, M.S.(2009) J Mol Biology 389: 863-879
- PubMed: 19394346
- DOI: https://doi.org/10.1016/j.jmb.2009.04.046
- Primary Citation of Related Structures:
3FSX, 3FSY - PubMed Abstract:
The enzyme tetrahydrodipicolinate N-succinyltransferase (DapD) is part of the L-lysine biosynthetic pathway. This pathway is crucial for the survival of the pathogen Mycobacterium tuberculosis (Mtb) and, consequently, the enzymes of the pathway are potential drug targets. We report here the crystal structures of Mtb-DapD and of Mtb-DapD in complex with the co-factor succinyl-CoA (SCoA) at 2.15 A and 1.97 A resolution, respectively. Each subunit of the trimeric enzyme consists of three domains, of which the second, a left-handed, parallel beta-helix (LbetaH domain), is the common structural motif of enzymes belonging to the hexapeptide repeat superfamily. The trimeric quaternary structure is stabilized by Mg(2+) and Na(+) located on the 3-fold axis. The binary complex of Mtb-DapD and SCoA reveals the binding mode(s) of the co-factor and a possible covalent reaction intermediate. The N-terminal domain of Mtb-DapD exhibits a unique architecture, including an interior water-filled channel, which allows access to a magnesium ion located at the 3-fold symmetry axis.
- EMBL Hamburg Outstation, Germany.
Organizational Affiliation: