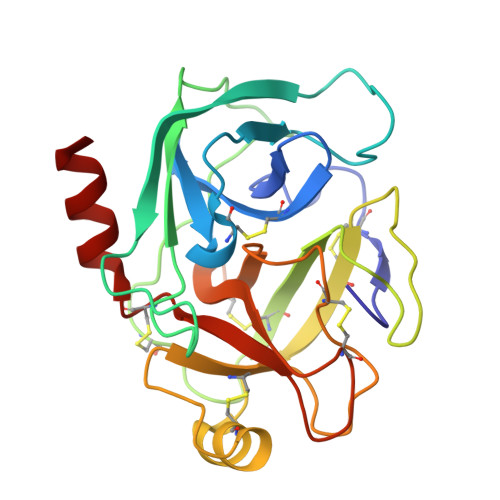
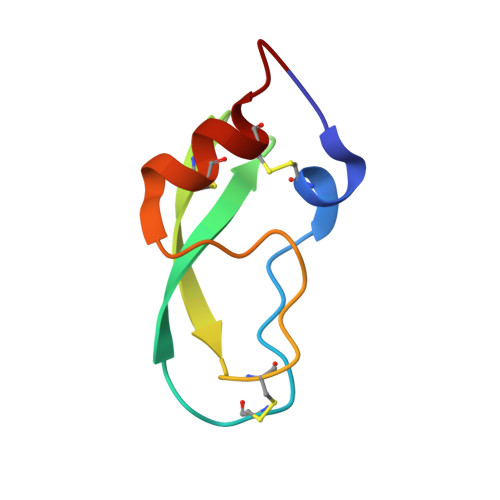
Structure of a serine protease poised to resynthesize a peptide bond.
Zakharova, E., Horvath, M.P., Goldenberg, D.P.(2009) Proc Natl Acad Sci U S A 106: 11034-11039
- PubMed: 19549826
- DOI: https://doi.org/10.1073/pnas.0902463106
- Primary Citation of Related Structures:
3FP6, 3FP7, 3FP8 - PubMed Abstract:
The serine proteases are among the most thoroughly studied enzymes, and numerous crystal structures representing the enzyme-substrate complex and intermediates in the hydrolysis reactions have been reported. Some aspects of the catalytic mechanism remain controversial, however, especially the role of conformational changes in the reaction. We describe here a high-resolution (1.46 A) crystal structure of a complex formed between a cleaved form of bovine pancreatic trypsin inhibitor (BPTI) and a catalytically inactive trypsin variant with the BPTI cleavage site ideally positioned in the active site for resynthesis of the peptide bond. This structure defines the positions of the newly generated amino and carboxyl groups following the 2 steps in the hydrolytic reaction. Comparison of this structure with those representing other intermediates in the reaction demonstrates that the residues of the catalytic triad are positioned to promote each step of both the forward and reverse reaction with remarkably little motion and with conservation of hydrogen-bonding interactions. The results also provide insights into the mechanism by which inhibitors like BPTI normally resist hydrolysis when bound to their target proteases.
- Department of Biology, University of Utah, Salt Lake City, UT 84112-0840, USA.
Organizational Affiliation: