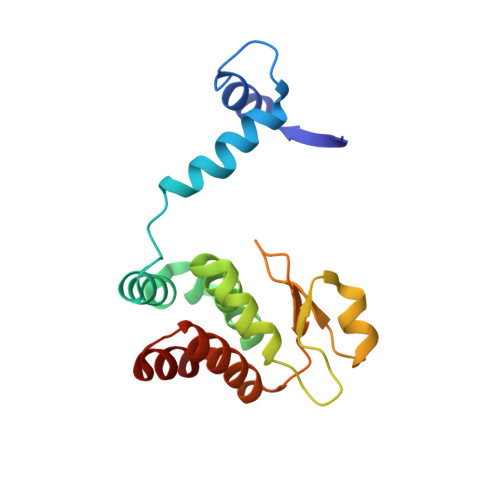
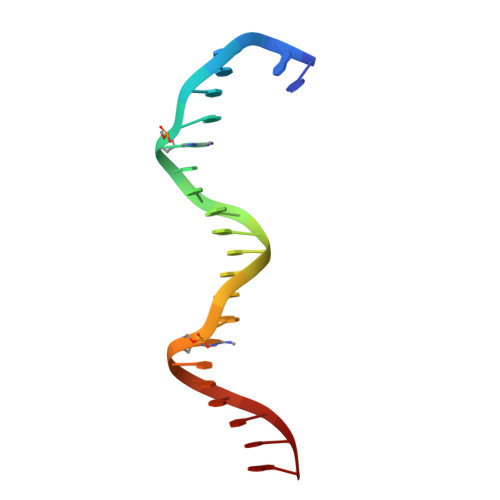
Structural insights into the cooperative binding of SeqA to a tandem GATC repeat
Chung, Y.S., Brendler, T., Austin, S., Guarne, A.(2009) Nucleic Acids Res 37: 3143-3152
- PubMed: 19304745
- DOI: https://doi.org/10.1093/nar/gkp151
- Primary Citation of Related Structures:
3FMT - PubMed Abstract:
SeqA is a negative regulator of DNA replication in Escherichia coli and related bacteria that functions by sequestering the origin of replication and facilitating its resetting after every initiation event. Inactivation of the seqA gene leads to unsynchronized rounds of replication, abnormal localization of nucleoids and increased negative superhelicity. Excess SeqA also disrupts replication synchrony and affects cell division. SeqA exerts its functions by binding clusters of transiently hemimethylated GATC sequences generated during replication. However, the molecular mechanisms that trigger formation and disassembly of such complex are unclear. We present here the crystal structure of a dimeric mutant of SeqA [SeqADelta(41-59)-A25R] bound to tandem hemimethylated GATC sites. The structure delineates how SeqA forms a high-affinity complex with DNA and it suggests why SeqA only recognizes GATC sites at certain spacings. The SeqA-DNA complex also unveils additional protein-protein interaction surfaces that mediate the formation of higher ordered complexes upon binding to newly replicated DNA. Based on this data, we propose a model describing how SeqA interacts with newly replicated DNA within the origin of replication and at the replication forks.
- Department of Biochemistry and Biomedical Sciences, Health Sciences Center, McMaster University, 1200 Main Street West, Hamilton, ON L8N 3Z5, Canada.
Organizational Affiliation: