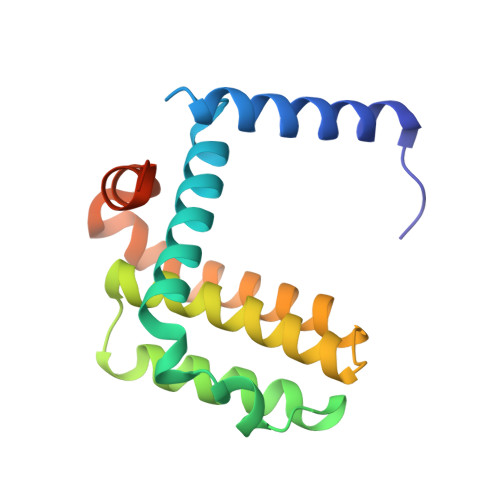
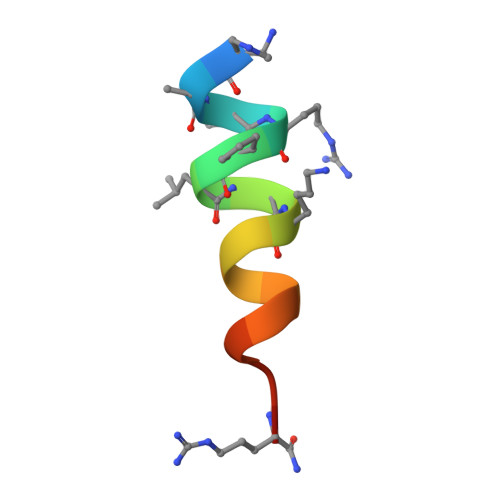
High-Resolution Structural Characterization of a Helical alpha/beta-Peptide Foldamer Bound to the Anti-Apoptotic Protein Bcl-x(L)
Lee, E.F., Sadowsky, J.D., Smith, B.J., Czabotar, P.E., Peterson-Kaufman, K.J., Colman, P.M., Gellman, S.H., Fairlie, W.D.(2009) Angew Chem Int Ed Engl 48: 4318-4322
- PubMed: 19229915
- DOI: https://doi.org/10.1002/anie.200805761
- Primary Citation of Related Structures:
3FDL, 3FDM - PubMed Abstract:
Get into the groove: The first high-resolution structure of a foldamer bound to a protein target is described (see picture; foldamer in sticks). The foldamer consists of alpha- and beta-amino acid residues and is bound to the anti-apoptotic protein Bcl-x(L). The overall binding mode and key interactions observed in the foldamer/Bcl-x(L) complex mimic those seen in complexes of Bcl-x(L) with natural alpha-peptide ligands. Additional contacts in the foldamer/Bcl-x(L) complex involving beta-amino acid residues appear to contribute to binding affinity.
- Structural Biology Division, The Walter and Eliza Hall Institute of Medical Research, 1G Royal Parade, Parkville, Victoria 3052, Australia.
Organizational Affiliation: