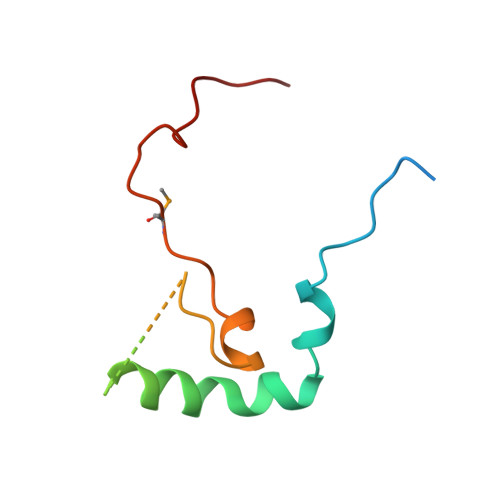
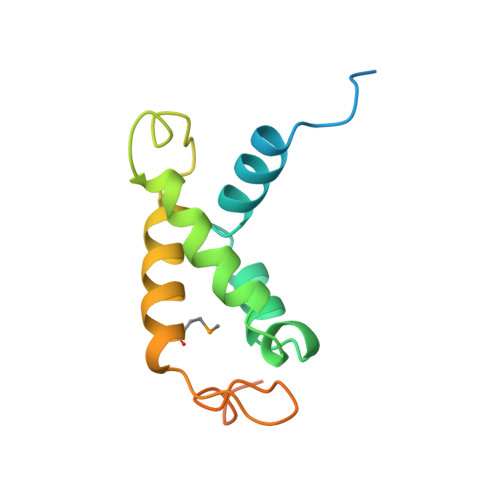
Identification, structure, and functional requirement of the Mediator submodule Med7N/31
Koschubs, T., Seizl, M., Lariviere, L., Kurth, F., Baumli, S., Martin, D.E., Cramer, P.(2009) EMBO J 28: 69-80
- PubMed: 19057509
- DOI: https://doi.org/10.1038/emboj.2008.254
- Primary Citation of Related Structures:
3FBI, 3FBN - PubMed Abstract:
Mediator is a modular multiprotein complex required for regulated transcription by RNA polymerase (Pol) II. Here, we show that the middle module of the Mediator core contains a submodule of unique structure and function that comprises the N-terminal part of subunit Med7 (Med7N) and the highly conserved subunit Med31 (Soh1). The Med7N/31 submodule shows a conserved novel fold, with two proline-rich stretches in Med7N wrapping around the right-handed four-helix bundle of Med31. In vitro, Med7N/31 is required for activated transcription and can act in trans when added exogenously. In vivo, Med7N/31 has a predominantly positive function on the expression of a specific subset of genes, including genes involved in methionine metabolism and iron transport. Comparative phenotyping and transcriptome profiling identify specific and overlapping functions of different Mediator submodules.
- Gene Center and Center for Integrated Protein Science Munich (CIPSM), Department of Chemistry and Biochemistry, Ludwig-Maximilians-Universität (LMU) München, Munich, Germany.
Organizational Affiliation: