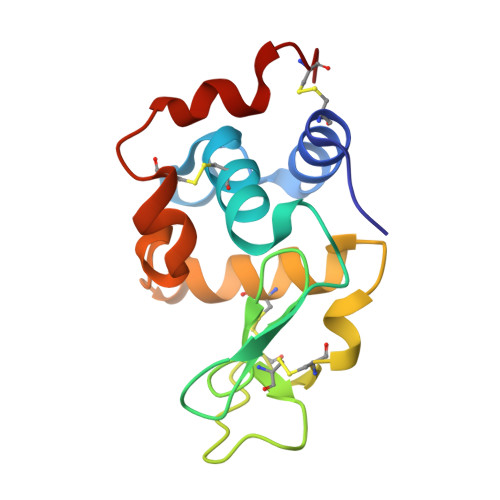
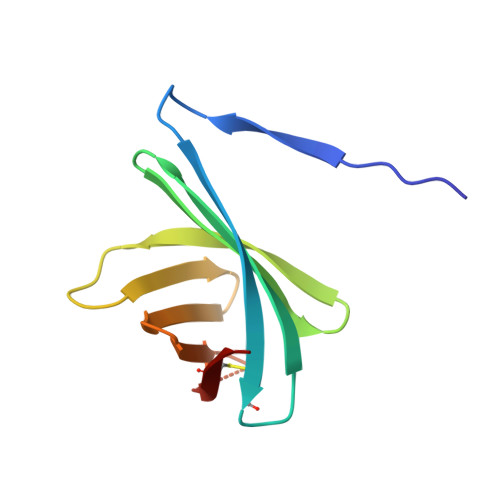
Structural basis for the recognition of lysozyme by MliC, a periplasmic lysozyme inhibitor in Gram-negative bacteria.
Yum, S., Kim, M.J., Xu, Y., Jin, X.L., Yoo, H.Y., Park, J.W., Gong, J.H., Choe, K.M., Lee, B.L., Ha, N.C.(2009) Biochem Biophys Res Commun 378: 244-248
- PubMed: 19028453
- DOI: https://doi.org/10.1016/j.bbrc.2008.11.039
- Primary Citation of Related Structures:
3F6Z - PubMed Abstract:
Lysozymes are an important component of the innate immune system of animals that hydrolyze peptidoglycan, the major bacterial cell wall constituent. Many bacteria have contrived various means of dealing with this bactericidal enzyme, one of which is to produce lysozyme inhibitors. Recently, a novel family of bacterial lysozyme inhibitors was identified in various Gram-negative bacteria, named MliC (membrane bound lysozyme inhibitor of C-type lysozyme). Here, we report the crystal structure of Pseudomonas aeruginosa MliC in complex with chicken egg white lysozyme. Combined with mutational study, the complex structure demonstrates that the invariant loop of MliC plays a crucial role in the inhibition of the lysozyme by its insertion to the active site cleft of the lysozyme, where the loop forms hydrogen and ionic bonds with the catalytic residues. Since MliC family members have been implicated as putative colonization or virulence factors, the structures and mechanism of action of MliC will be of relevance to the control of bacterial growth in animal hosts.
- College of Pharmacy and Research Institute for Drug Development, Pusan National University, Room #311, Pharmacy Building, Jangjeon-dong Geumjeong-gu, Busan 609-735, Republic of Korea.
Organizational Affiliation: