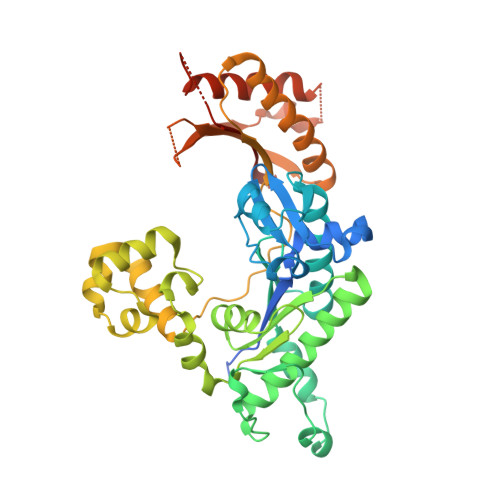
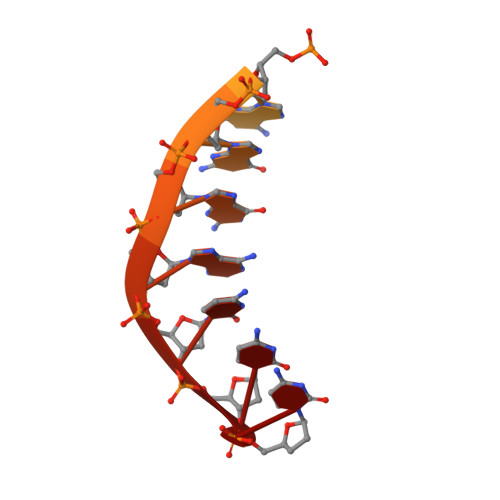
Lesion bypass of N2-ethylguanine by human DNA polymerase iota.
Pence, M.G., Blans, P., Zink, C.N., Hollis, T., Fishbein, J.C., Perrino, F.W.(2009) J Biological Chem 284: 1732-1740
- PubMed: 18984581
- DOI: https://doi.org/10.1074/jbc.M807296200
- Primary Citation of Related Structures:
3EPG, 3EPI - PubMed Abstract:
Nucleotide incorporation and extension opposite N2-ethyl-Gua by DNA polymerase iota was measured and structures of the DNA polymerase iota-N2-ethyl-Gua complex with incoming nucleotides were solved. Efficiency and fidelity of DNA polymerase iota opposite N2-ethyl-Gua was determined by steady state kinetic analysis with Mg2+ or Mn2+ as the activating metal. DNA polymerase iota incorporates dCMP opposite N2-ethyl-Gua and unadducted Gua with similar efficiencies in the presence of Mg2+ and with greater efficiencies in the presence of Mn2+. However, the fidelity of nucleotide incorporation by DNA polymerase iota opposite N2-ethyl-Gua and Gua using Mn2+ is lower relative to that using Mg2+ indicating a metal-dependent effect. DNA polymerase iota extends from the N2-ethyl-Gua:Cyt 3' terminus more efficiently than from the Gua:Cyt base pair. Together these kinetic data indicate that the DNA polymerase iota catalyzed reaction is well suited for N(2)-ethyl-Gua bypass. The structure of DNA polymerase iota with N2-ethyl-Gua at the active site reveals the adducted base in the syn configuration when the correct incoming nucleotide is present. Positioning of the ethyl adduct into the major groove removes potential steric overlap between the adducted template base and the incoming dCTP. Comparing structures of DNA polymerase iota complexed with N2-ethyl-Gua and Gua at the active site suggests movements in the DNA polymerase iota polymerase-associated domain to accommodate the adduct providing direct evidence that DNA polymerase iota efficiently replicates past a minor groove DNA adduct by positioning the adducted base in the syn configuration.
- Department of Biochemistry and Center for Structural Biology, Wake Forest University Health Sciences, Winston-Salem, North Carolina 27157, USA.
Organizational Affiliation: