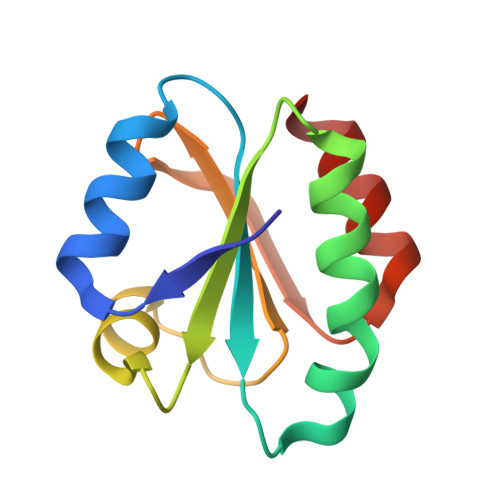
Structure of human thioredoxin exhibits a large conformational change.
Hall, G., Emsley, J.(2010) Protein Sci 19: 1807-1811
- PubMed: 20661909
- DOI: https://doi.org/10.1002/pro.466
- Primary Citation of Related Structures:
3E3E - PubMed Abstract:
Thioredoxin is an oxidoreductase, which is ubiquitously present across phyla from humans to plants and bacteria. Thioredoxin reduces a variety of substrates through active site Cys 32, which is subsequently oxidized to form the intramolecular disulphide with Cys 35. The thioredoxin fold is known to be highly stable and conformational changes in the active site loops and residues Cys 32, Cys 35 have been characterized between ligand bound and free structures. We have determined a novel 2.0 A resolution crystal structure for a human thioredoxin, which reveals a much larger conformational change than previously characterized. The principal change involves unraveling of a helix to form an extended loop that is linked to secondary changes in further loop regions and the wider area of the active site Cys 32. This gives rise to a more open conformation and an elongated hydrophobic pocket results in place of the helix. Buried residue Cys 62 from this helix becomes exposed in the open conformation. This provides a structural basis for observations that the Cys 62 sidechain can form mixed disulphides and be modified by thiol reactive small molecules.
- Centre for Biomolecular Sciences, School of Pharmacy, University of Nottingham, Nottingham, United Kingdom.
Organizational Affiliation: