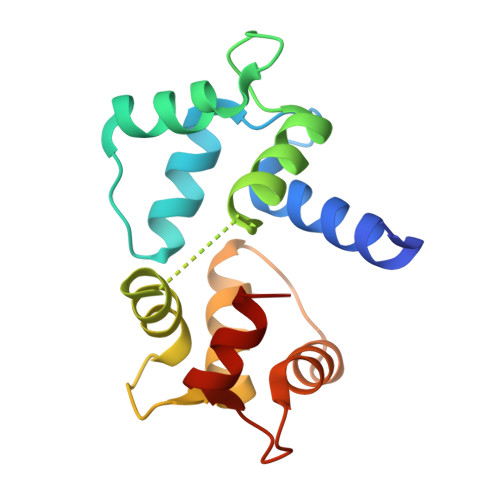
Structures of Ca(V)2 Ca(2+)/CaM-IQ Domain Complexes Reveal Binding Modes that Underlie Calcium-Dependent Inactivation and Facilitation.
Kim, E.Y., Rumpf, C.H., Fujiwara, Y., Cooley, E.S., Van Petegem, F., Minor, D.L.(2008) Structure 16: 1455-1467
- PubMed: 18940602
- DOI: https://doi.org/10.1016/j.str.2008.07.010
- Primary Citation of Related Structures:
3DVE, 3DVJ, 3DVK, 3DVM - PubMed Abstract:
Calcium influx drives two opposing voltage-activated calcium channel (Ca(V)) self-modulatory processes: calcium-dependent inactivation (CDI) and calcium-dependent facilitation (CDF). Specific Ca(2+)/calmodulin (Ca(2+)/CaM) lobes produce CDI and CDF through interactions with the Ca(V)alpha(1) subunit IQ domain. Curiously, Ca(2+)/CaM lobe modulation polarity appears inverted between Ca(V)1s and Ca(V)2s. Here, we present crystal structures of Ca(V)2.1, Ca(V)2.2, and Ca(V)2.3 Ca(2+)/CaM-IQ domain complexes. All display binding orientations opposite to Ca(V)1.2 with a physical reversal of the CaM lobe positions relative to the IQ alpha-helix. Titration calorimetry reveals lobe competition for a high-affinity site common to Ca(V)1 and Ca(V)2 IQ domains that is occupied by the CDI lobe in the structures. Electrophysiological experiments demonstrate that the N-terminal Ca(V)2 Ca(2+)/C-lobe anchors affect CDF. Together, the data unveil the remarkable structural plasticity at the heart of Ca(V) feedback modulation and indicate that Ca(V)1 and Ca(V)2 IQ domains bear a dedicated CDF site that exchanges Ca(2+)/CaM lobe occupants.
- Cardiovascular Research Institute, Department of Biochemistry and Biophysics, California Institute for Quantitative Biosciences, University of California, San Francisco, CA 94158-2330, USA.
Organizational Affiliation: