Structural basis for group A trichothiodystrophy
Kainov, D.E., Vitorino, M., Cavarelli, J., Poterszman, A., Egly, J.M.(2008) Nat Struct Mol Biol 15: 980-984
- PubMed: 19172752
- DOI: https://doi.org/10.1038/nsmb.1478
- Primary Citation of Related Structures:
3DGP, 3DOM - PubMed Abstract:
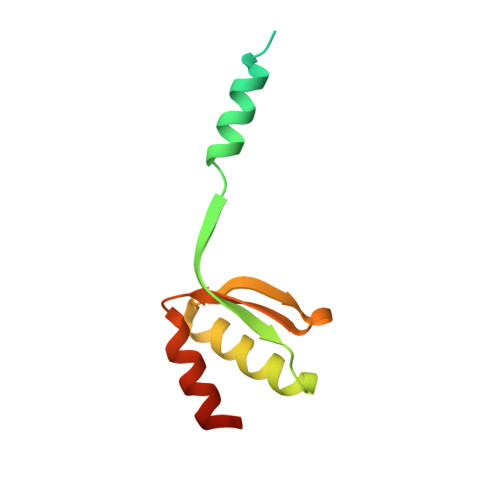
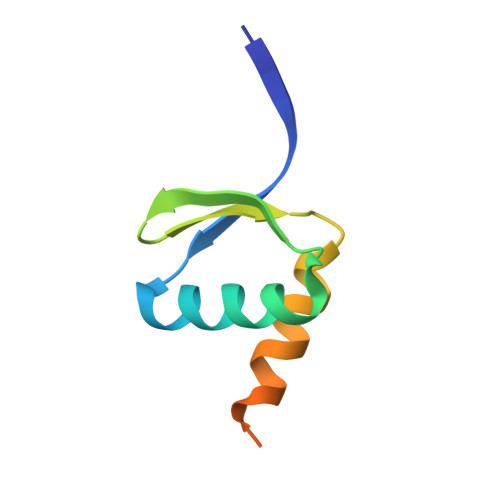
Patients with the rare neurodevelopmental repair syndrome known as group A trichothiodystrophy (TTD-A) carry mutations in the gene encoding the p8 subunit of the transcription and DNA repair factor TFIIH. Here we describe the crystal structure of a minimal complex between Tfb5, the yeast ortholog of p8, and the C-terminal domain of Tfb2, the yeast p52 subunit of TFIIH. The structure revealed that these two polypeptides adopt the same fold, forming a compact pseudosymmetric heterodimer via a beta-strand addition and coiled coils interactions between terminal alpha-helices. Furthermore, Tfb5 protects a hydrophobic surface in Tfb2 from solvent, providing a rationale for the influence of p8 in the stabilization of p52 and explaining why mutations that weaken p8-p52 interactions lead to a reduced intracellular TFIIH concentration and a defect in nucleotide-excision repair, a common feature of TTD cells.
- lnstitut de Génétique et de Biologie Moléculaire et Cellulaire, CNRS/INSERM/ULP, BP 163, 67404 Illkirch Cedex, France.
Organizational Affiliation: