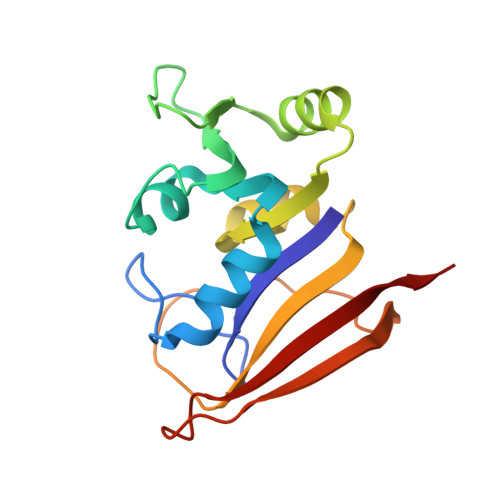
Crystal structures of Escherichia coli and Lactobacillus casei dihydrofolate reductase refined at 1.7 A resolution. I. General features and binding of methotrexate.
Bolin, J.T., Filman, D.J., Matthews, D.A., Hamlin, R.C., Kraut, J.(1982) J Biological Chem 257: 13650-13662
- PubMed: 6815178
- Primary Citation of Related Structures:
3DFR, 4DFR - PubMed Abstract:
X-ray data have been extended to 1.7 A for a binary complex of Escherichia coli dihydrofolate reductase with methotrexate and a ternary complex of Lactobacillus casei dihydrofolate reductase with methotrexate and NADPH. Models for both structures have been refined to R factors of 0.15 and include parameters for fixed and liquid solvent. The two species of dihydrofolate reductase resemble one another even more closely than was thought to be the case prior to refinement. Several new structural features have also been discovered. Among them are a cis peptide linking Gly-97 and Gly-98 (L. Casei numbering) in both species, an alpha helix involving residues 43 through 50 in the E. coli enzyme, and the existence of what may be a specific hydration site on exposed alpha helices. Refinement has led to a revised description of the details of methotrexate binding. We now see that a fixed water molecule mediates the interaction between methotrexate's 2-amino group and Thr-116 (L. casei numbering) and that the inhibitor's 4-amino group makes two hydrogen bonds with the enzyme (instead of one). Other revisions are also discussed. A hypothetical model for substrate binding is proposed in which the pteridine ring is turned upside down while all protein and solvent atoms remain fixed. Asp-26 in this model is hydrogen bonded to the substrate's 2-amino group and to N3.