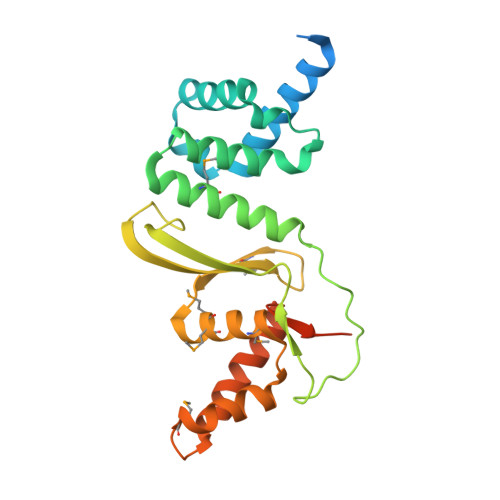
The structure of the first representative of Pfam family PF09836 reveals a two-domain organization and suggests involvement in transcriptional regulation.
Das, D., Grishin, N.V., Kumar, A., Carlton, D., Bakolitsa, C., Miller, M.D., Abdubek, P., Astakhova, T., Axelrod, H.L., Burra, P., Chen, C., Chiu, H.J., Chiu, M., Clayton, T., Deller, M.C., Duan, L., Ellrott, K., Ernst, D., Farr, C.L., Feuerhelm, J., Grzechnik, A., Grzechnik, S.K., Grant, J.C., Han, G.W., Jaroszewski, L., Jin, K.K., Johnson, H.A., Klock, H.E., Knuth, M.W., Kozbial, P., Krishna, S.S., Marciano, D., McMullan, D., Morse, A.T., Nigoghossian, E., Nopakun, A., Okach, L., Oommachen, S., Paulsen, J., Puckett, C., Reyes, R., Rife, C.L., Sefcovic, N., Tien, H.J., Trame, C.B., van den Bedem, H., Weekes, D., Wooten, T., Xu, Q., Hodgson, K.O., Wooley, J., Elsliger, M.A., Deacon, A.M., Godzik, A., Lesley, S.A., Wilson, I.A.(2010) Acta Crystallogr Sect F Struct Biol Cryst Commun 66: 1174-1181
- PubMed: 20944208
- DOI: https://doi.org/10.1107/S1744309109022672
- Primary Citation of Related Structures:
3DEE - PubMed Abstract:
Proteins with the DUF2063 domain constitute a new Pfam family, PF09836. The crystal structure of a member of this family, NGO1945 from Neisseria gonorrhoeae, has been determined and reveals that the N-terminal DUF2063 domain is likely to be a DNA-binding domain. In conjunction with the rest of the protein, NGO1945 is likely to be involved in transcriptional regulation, which is consistent with genomic neighborhood analysis. Of the 216 currently known proteins that contain a DUF2063 domain, the most significant sequence homologs of NGO1945 (∼40-99% sequence identity) are from various Neisseria and Haemophilus species. As these are important human pathogens, NGO1945 represents an interesting candidate for further exploration via biochemical studies and possible therapeutic intervention.
- Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, Menlo Park, CA, USA.
Organizational Affiliation: