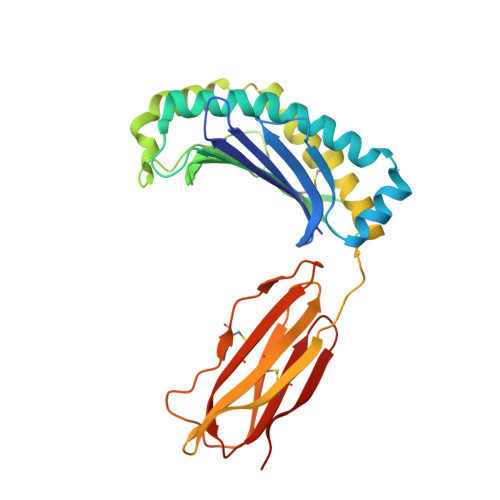
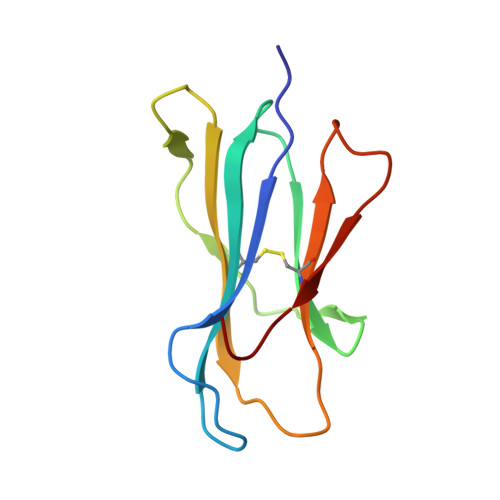
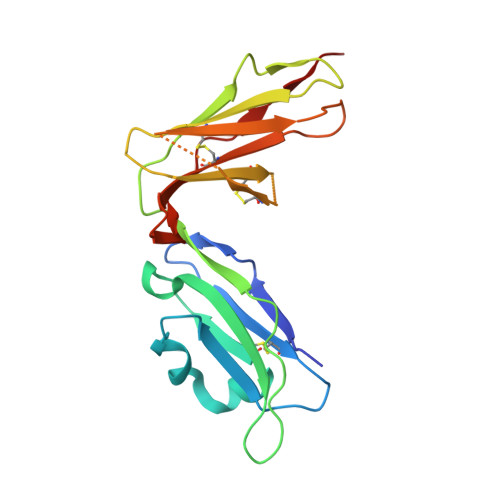
Structure of UL18, a peptide-binding viral MHC mimic, bound to a host inhibitory receptor
Yang, Z., Bjorkman, P.J.(2008) Proc Natl Acad Sci U S A 105: 10095-10100
- PubMed: 18632577
- DOI: https://doi.org/10.1073/pnas.0804551105
- Primary Citation of Related Structures:
3D2U - PubMed Abstract:
UL18 is a human cytomegalovirus class I MHC (MHCI) homolog that binds the host inhibitory receptor LIR-1 and the only known viral MHC homolog that presents peptides. The 2.2-A structure of a LIR-1/UL18/peptide complex reveals increased contacts and optimal surface complementarity in the LIR-1/UL18 interface compared with LIR/MHCI interfaces, resulting in a >1,000-fold higher affinity. Despite sharing only approximately 25% sequence identity, UL18's structure and peptide binding are surprisingly similar to host MHCI. The crystal structure suggests that most of the UL18 surface, except where LIR-1 and the host-derived light chain bind, is covered by carbohydrates attached to 13 potential N-glycosylation sites, thereby preventing access to bound peptide and association with most MHCI-binding proteins. The LIR-1/UL18 structure demonstrates how a viral protein evolves from its host ancestor to impede unwanted interactions while preserving and improving its receptor-binding site.
- Division of Biology 114-96 and Howard Hughes Medical Institute, California Institute of Technology, Pasadena, CA 91125, USA.
Organizational Affiliation: