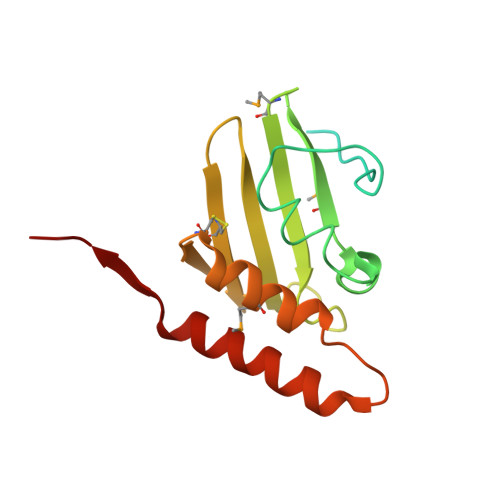
The Helicobacter pylori CagD (HP0545, Cag24) protein is essential for CagA translocation and maximal induction of interleukin-8 secretion.
Cendron, L., Couturier, M., Angelini, A., Barison, N., Stein, M., Zanotti, G.(2009) J Mol Biology 386: 204-217
- PubMed: 19109970
- DOI: https://doi.org/10.1016/j.jmb.2008.12.018
- Primary Citation of Related Structures:
3CWX, 3CWY - PubMed Abstract:
Pathogenic strains of Helicobacter pylori use a type IV secretion system (T4SS) to deliver the toxin CagA into human host cells. The T4SS, along with the toxin itself, is coded into a genomic insert, which is termed the cag pathogenicity island. The cag pathogenicity island contains about 30 open-reading frames, for most of which the exact function is not well characterized or totally unknown. We have determined the crystal structure of one of the proteins coded by the cag genes, CagD, in two crystal forms. We show that the protein is a covalent dimer in which each monomer folds as a single domain that is composed of five beta-strands and three alpha-helices. Our data show that in addition to a cytosolic pool, CagD partially associates with the inner membrane, where it may be exposed to the periplasmic space. Furthermore, CagA tyrosine phosphorylation and interleukin-8 assays identified CagD as a crucial component of the T4SS that is involved in CagA translocation into host epithelial cells; however, it does not seem absolutely necessary for pilus assembly. We have also identified significant amounts of CagD in culture supernatants, which are not a result of general bacterial lysis. Since this localization was independent of the various tested cag mutants, our findings may indicate that CagD is released into the supernatant during host cell infection and then binds to the host cell surface or is incorporated in the pilus structure. Overall, our results suggest that CagD may serve as a unique multifunctional component of the T4SS that may be involved in CagA secretion at the inner membrane and may localize outside the bacteria to promote additional effects on the host cell.
- Department of Chemistry, University of Padua Via Marzolo 1, I-35131 Padua, Italy.
Organizational Affiliation: