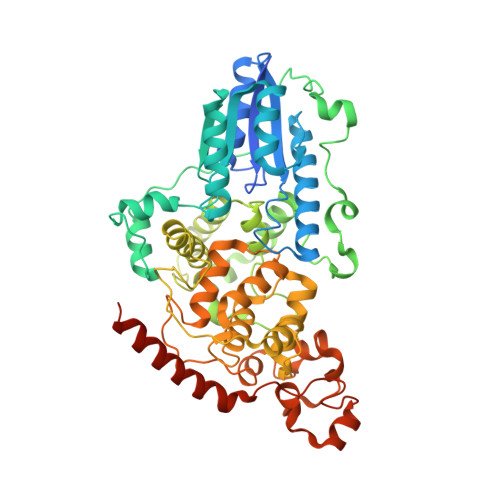
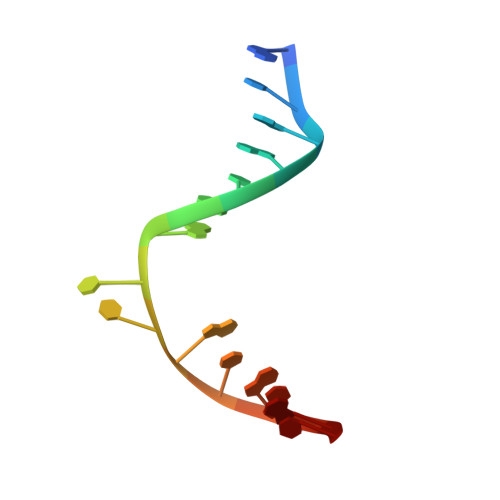
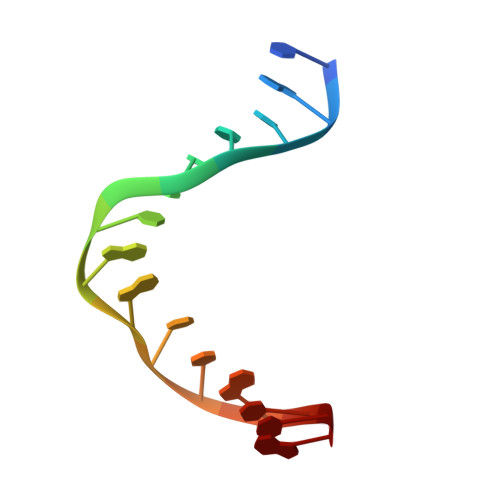
Crystal structure and mechanism of a DNA (6-4) photolyase.
Maul, M.J., Barends, T.R., Glas, A.F., Cryle, M.J., Domratcheva, T., Schneider, S., Schlichting, I., Carell, T.(2008) Angew Chem Int Ed Engl 47: 10076-10080
- PubMed: 18956392
- DOI: https://doi.org/10.1002/anie.200804268
- Primary Citation of Related Structures:
3CVU, 3CVY - Centre for Integrative Protein Science, Department of Chemistry and Biochemistry, Ludwig-Maximilians University Munich, Butenandtstrasse 5-13, 81377 Munich, Germany.
Organizational Affiliation: