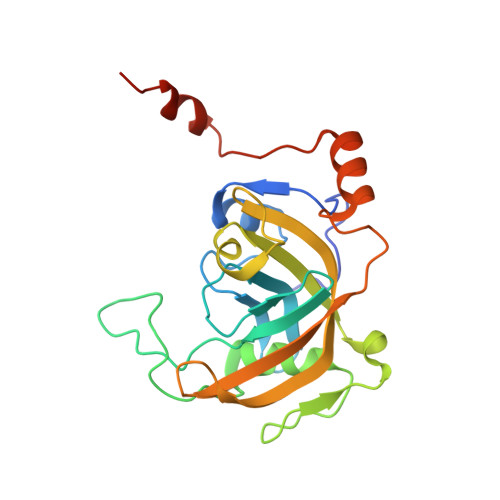
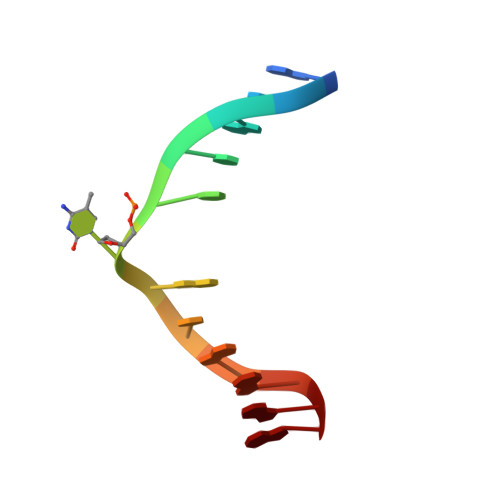
Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1.
Avvakumov, G.V., Walker, J.R., Xue, S., Li, Y., Duan, S., Bronner, C., Arrowsmith, C.H., Dhe-Paganon, S.(2008) Nature 455: 822-825
- PubMed: 18772889
- DOI: https://doi.org/10.1038/nature07273
- Primary Citation of Related Structures:
3BI7, 3CLZ - PubMed Abstract:
Epigenetic inheritance in mammals is characterized by high-fidelity replication of CpG methylation patterns during development. UHRF1 (also known as ICBP90 in humans and Np95 in mouse) is an E3 ligase important for the maintenance of global and local DNA methylation in vivo. The preferential affinity of UHRF1 for hemi-methylated DNA over symmetrically methylated DNA by means of its SET and RING-associated (SRA) domain and its association with the maintenance DNA methyltransferase 1 (DNMT1) suggests a role in replication of the epigenetic code. Here we report the 1.7 A crystal structure of the apo SRA domain of human UHRF1 and a 2.2 A structure of its complex with hemi-methylated DNA, revealing a previously unknown reading mechanism for methylated CpG sites (mCpG). The SRA-DNA complex has several notable structural features including a binding pocket that accommodates the 5-methylcytosine that is flipped out of the duplex DNA. Two specialized loops reach through the resulting gap in the DNA from both the major and the minor grooves to read the other three bases of the CpG duplex. The major groove loop confers both specificity for the CpG dinucleotide and discrimination against methylation of deoxycytidine of the complementary strand. The structure, along with mutagenesis data, suggests how UHRF1 acts as a key factor for DNMT1 maintenance methylation through recognition of a fundamental unit of epigenetic inheritance, mCpG.
- Structural Genomics Consortium, University of Toronto, 100 College Street, Toronto, Ontario M5G 1L5, Canada.
Organizational Affiliation: