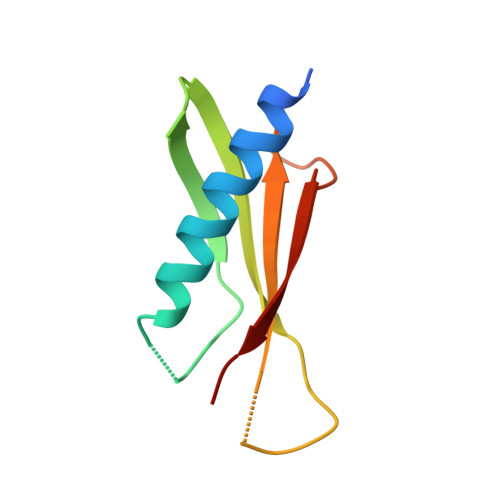
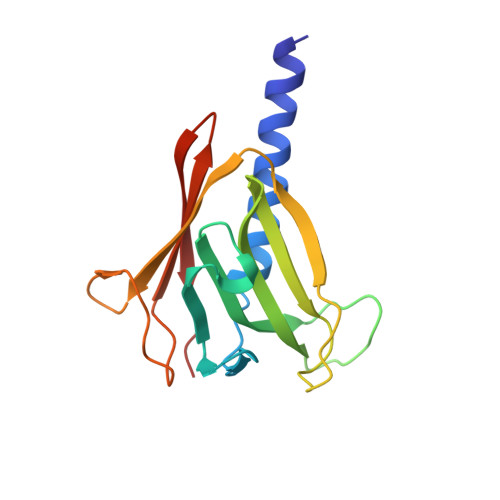
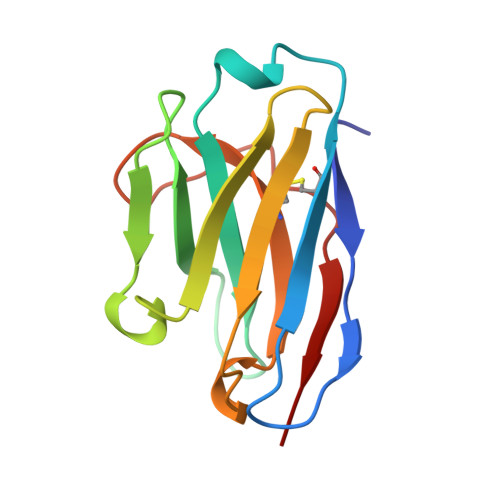
Nanobody-aided structure determination of the EpsI:EpsJ pseudopilin heterodimer from Vibrio vulnificus.
Lam, A.Y., Pardon, E., Korotkov, K.V., Hol, W.G., Steyaert, J.(2009) J Struct Biol 166: 8-15
- PubMed: 19118632
- DOI: https://doi.org/10.1016/j.jsb.2008.11.008
- Primary Citation of Related Structures:
3CFI - PubMed Abstract:
Pseudopilins form the central pseudopilus of the sophisticated bacterial type 2 secretion systems. The crystallization of the EpsI:EpsJ pseudopilin heterodimer from Vibrio vulnificus was greatly accelerated by the use of nanobodies, which are the smallest antigen-binding fragments derived from heavy-chain only camelid antibodies. Seven anti-EpsI:EpsJ nanobodies were generated and co-crystallization of EpsI:EpsJ nanobody complexes yielded several crystal forms very rapidly. In the structure solved, the nanobodies are arranged in planes throughout the crystal lattice, linking layers of EpsI:EpsJ heterodimers. The EpsI:EpsJ dimer observed confirms a right-handed architecture of the pseudopilus, but, compared to a previous structure of the EpsI:EpsJ heterodimer, EpsI differs 6 degrees in orientation with respect to EpsJ; one loop of EpsJ is shifted by approximately 5A due to interactions with the nanobody; and a second loop of EpsJ underwent a major change of 17A without contacts with the nanobody. Clearly, nanobodies accelerate dramatically the crystallization of recalcitrant protein complexes and can reveal conformational flexibility not observed before.
- Department of Biochemistry, Biomolecular Structure Center, University of Washington, 1959 Pacific Ave. NE, HSC K-428, Seattle, WA 98195, USA.
Organizational Affiliation: