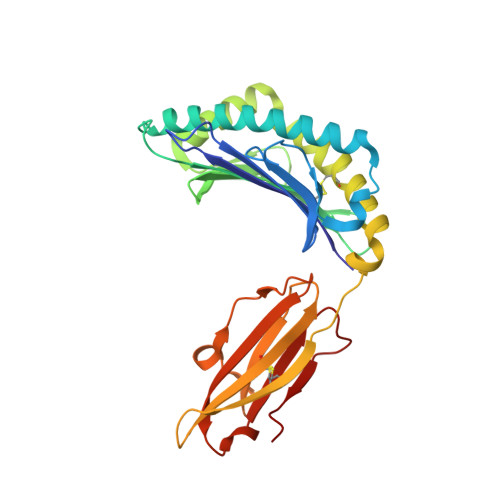
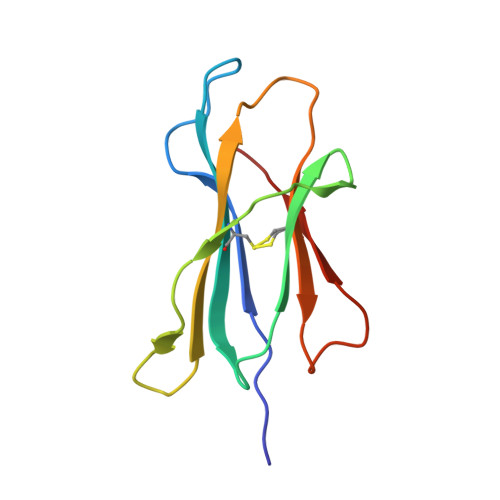
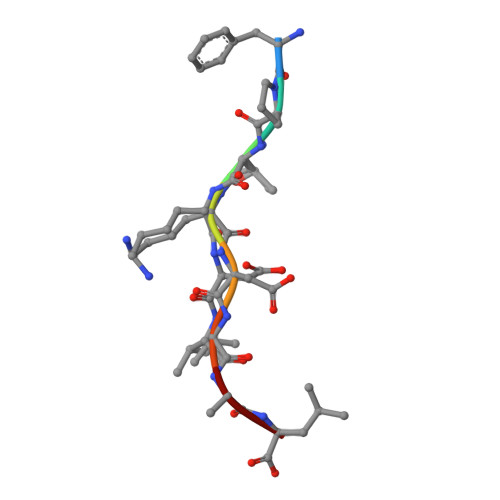
Impact of clonal competition for peptide-MHC complexes on the CD8+ T-cell repertoire selection in a persistent viral infection
Wynn, K.K., Fulton, Z., Cooper, L., Silins, S.L., Gras, S., Archbold, J.K., Tynan, F.E., Miles, J.J., McCluskey, J., Burrows, S.R., Rossjohn, J., Khanna, R.(2008) Blood 111: 4283-4292
- PubMed: 18270323
- DOI: https://doi.org/10.1182/blood-2007-11-122622
- Primary Citation of Related Structures:
3BW9, 3BWA - PubMed Abstract:
CD8(+) T-cell responses to persistent viral infections are characterized by the accumulation of an oligoclonal T-cell repertoire and a reduction in the naive T-cell pool. However, the precise mechanism for this phenomenon remains elusive. Here we show that human cytomegalovirus (HCMV)-specific CD8(+) T cells recognizing distinct epitopes from the pp65 protein and restricted through an identical HLA class I allele (HLA B*3508) exhibited either a highly conserved public T-cell repertoire or a private, diverse T-cell response, which was uniquely altered in each donor following in vitro antigen exposure. Selection of a public T-cell receptor (TCR) was coincident with an atypical major histocompatibility complex (MHC)-peptide structure, in that the epitope adopted a helical conformation that bulged from the peptide-binding groove, while a diverse TCR profile was observed in response to the epitope that formed a flatter, more "featureless" landscape. Clonotypes with biased TCR usage demonstrated more efficient recognition of virus-infected cells, a greater CD8 dependency, and were more terminally differentiated in their phenotype when compared with the T cells expressing diverse TCR. These findings provide new insights into our understanding on how the biology of antigen presentation in addition to the structural features of the pMHC-I might shape the T-cell repertoire and its phenotype.
- Australian Centre for Vaccine Development and Division of Infectious Diseases and Immunology, Queensland Institute of Medical Research, Brisbane, Australia.
Organizational Affiliation: