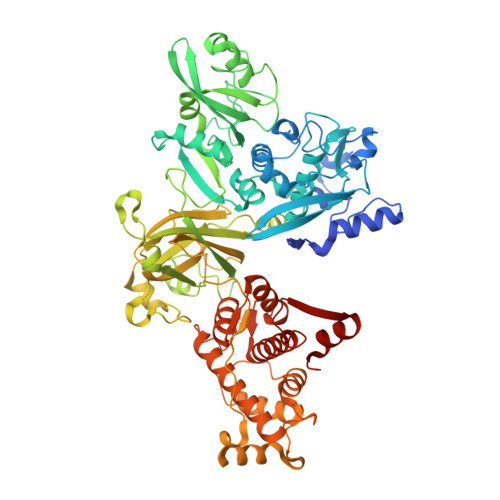
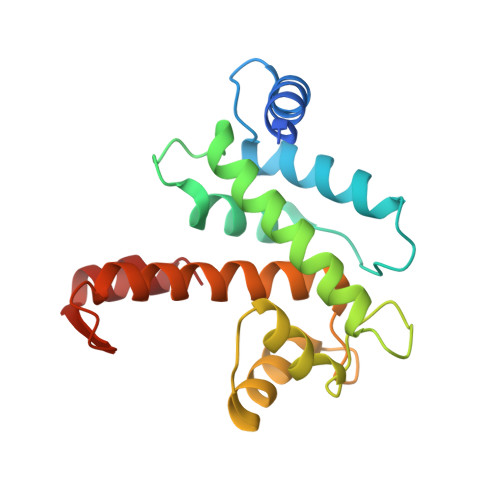
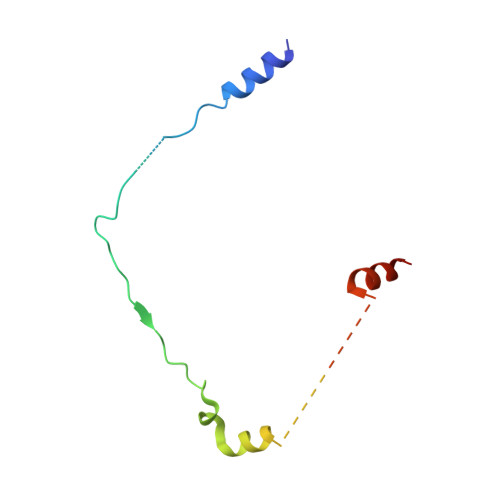
Calcium-bound structure of calpain and its mechanism of inhibition by calpastatin.
Hanna, R.A., Campbell, R.L., Davies, P.L.(2008) Nature 456: 409-412
- PubMed: 19020623
- DOI: https://doi.org/10.1038/nature07451
- Primary Citation of Related Structures:
3BOW - PubMed Abstract:
Calpains are non-lysosomal calcium-dependent cysteine proteinases that selectively cleave proteins in response to calcium signals and thereby control cellular functions such as cytoskeletal remodelling, cell cycle progression, gene expression and apoptotic cell death. In mammals, the two best-characterized members of the calpain family, calpain 1 and calpain 2 (micro-calpain and m-calpain, respectively), are ubiquitously expressed. The activity of calpains is tightly controlled by the endogenous inhibitor calpastatin, which is an intrinsically unstructured protein capable of reversibly binding and inhibiting four molecules of calpain, but only in the presence of calcium. To date, the mechanism of inhibition by calpastatin and the basis for its absolute specificity have remained speculative. It was not clear how this unstructured protein inhibits calpains without being cleaved itself, nor was it known how calcium induced changes that facilitated the binding of calpastatin to calpain. Here we report the 2.4-A-resolution crystal structure of the calcium-bound calpain 2 heterodimer bound by one of the four inhibitory domains of calpastatin. Calpastatin is seen to inhibit calpain by occupying both sides of the active site cleft. Although the inhibitor passes through the active site cleft it escapes cleavage in a novel manner by looping out and around the active site cysteine. The inhibitory domain of calpastatin recognizes multiple lower affinity sites present only in the calcium-bound form of the enzyme, resulting in an interaction that is tight, specific and calcium dependent. This crystal structure, and that of a related complex, also reveal the conformational changes that calpain undergoes on binding calcium, which include opening of the active site cleft and movement of the domains relative to each other to produce a more compact enzyme.
- Department of Biochemistry, Queen's University, Kingston, Ontario, Canada K7L 3N6.
Organizational Affiliation: