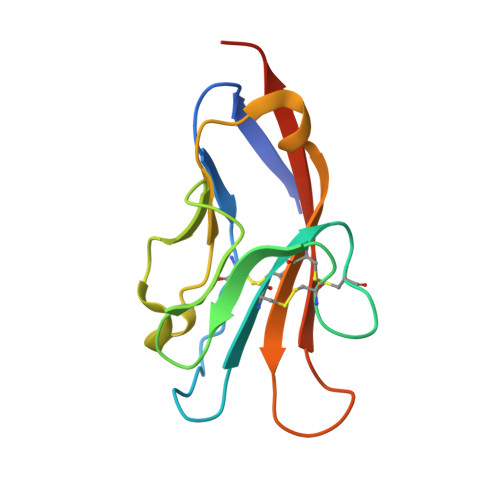
Structures of T Cell Immunoglobulin Mucin Protein 4 Show a Metal-Ion-Dependent Ligand Binding Site where Phosphatidylserine Binds.
Santiago, C., Ballesteros, A., Martinez-Munoz, L., Mellado, M., Kaplan, G.G., Freeman, G.J., Casasnovas, J.M.(2007) Immunity 27: 941-951
- PubMed: 18083575
- DOI: https://doi.org/10.1016/j.immuni.2007.11.008
- Primary Citation of Related Structures:
3BI9, 3BIA, 3BIB - PubMed Abstract:
The T cell immunoglobulin and mucin domain (TIM) proteins are important regulators of T cell responses. Crystal structures of the murine TIM-4 identified a metal-ion-dependent ligand binding site (MILIBS) in the immunoglobulin (Ig) domain of the TIM family. The characteristic CC' loop of the TIM domain and the hydrophobic FG loop shaped a narrow cavity where acidic compounds penetrate and coordinate to a metal ion bound to conserved residues in the TIM proteins. The structure of phosphatidylserine bound to the Ig domain showed that the hydrophilic head penetrates into the MILIBS and coordinates with the metal ion, whereas the aromatic residues on the tip of the FG loop interacted with the fatty acid chains and could insert into the lipid bilayer. Our results also revealed an important role of the MILIBS in the trafficking of TIM-1 to the cell surface.
- Centro Nacional de Biotecnología, CSIC, Campus Universidad Autónoma, 28049 Madrid, Spain.
Organizational Affiliation: