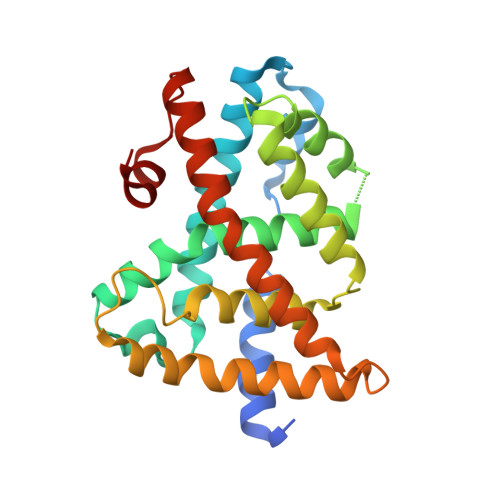
Identification of a potent synthetic FXR agonist with an unexpected mode of binding and activation.
Soisson, S.M., Parthasarathy, G., Adams, A.D., Sahoo, S., Sitlani, A., Sparrow, C., Cui, J., Becker, J.W.(2008) Proc Natl Acad Sci U S A 105: 5337-5342
- PubMed: 18391212
- DOI: https://doi.org/10.1073/pnas.0710981105
- Primary Citation of Related Structures:
3BEJ - PubMed Abstract:
The farnesoid X receptor (FXR), a member of the nuclear hormone receptor family, plays important roles in the regulation of bile acid and cholesterol homeostasis, glucose metabolism, and insulin sensitivity. There is intense interest in understanding the mechanisms of FXR regulation and in developing pharmaceutically suitable synthetic FXR ligands that might be used to treat metabolic syndrome. We report here the identification of a potent FXR agonist (MFA-1) and the elucidation of the structure of this ligand in ternary complex with the human receptor and a coactivator peptide fragment using x-ray crystallography at 1.9-A resolution. The steroid ring system of MFA-1 binds with its D ring-facing helix 12 (AF-2) in a manner reminiscent of hormone binding to classical steroid hormone receptors and the reverse of the pose adopted by naturally occurring bile acids when bound to FXR. This binding mode appears to be driven by the presence of a carboxylate on MFA-1 that is situated to make a salt-bridge interaction with an arginine residue in the FXR-binding pocket that is normally used to neutralize bound bile acids. Receptor activation by MFA-1 differs from that by bile acids in that it relies on direct interactions between the ligand and residues in helices 11 and 12 and only indirectly involves a protonated histidine that is part of the activation trigger. The structure of the FXR:MFA-1 complex differs significantly from that of the complex with a structurally distinct agonist, fexaramine, highlighting the inherent plasticity of the receptor.
- Merck Research Laboratories, POB 2000, Rahway, NJ 07065, USA. stephen_soisson@merck.com
Organizational Affiliation: