Molecular insights into miRNA processing by Arabidopsis thaliana SERRATE
Machida, S., Chen, H.Y., Yuan, Y.A.(2011) Nucleic Acids Res 39: 7828-7836
- PubMed: 21685453
- DOI: https://doi.org/10.1093/nar/gkr428
- Primary Citation of Related Structures:
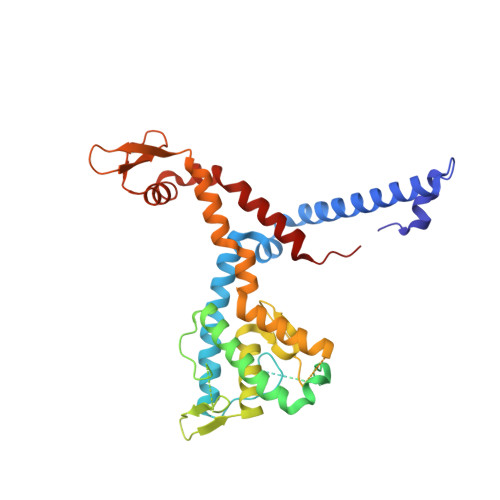
3AX1 - PubMed Abstract:
In plant, primary transcripts (pri-miRNAs) transcribed from miRNA genes by RNA polymerase II are first processed into stem-loop pre-miRNAs and further chopped into ∼21 nt long miRNAs by RNase III-like enzyme DCL1. SERRATE (SE) protein is an essential component for miRNA processing by assisting DCL1 for accurate cleavage. Here we report the crystal structure of Arabidopsis SE core (residues 194-543) at 2.7 Å. SE core adopts the 'walking man-like' topology with N-terminal α helices, C-terminal non-canonical zinc-finger domain and novel Middle domain resembling the leading leg, the lagging leg and the body, respectively. Pull-down assay shows that SE core provides the platform for HYL1 and DCL1 binding, whereas in vitro RNA binding and in vivo mutant rescue experiments suggest that the non-canonical zinc-finger domain coupled with C-terminal tail binds miRNA precursors. SE presumably works as a scaffold-like protein capable of binding both protein and RNA to guide the positioning of miRNA precursor toward DCL1 catalytic site within miRNA processing machinery in plant.
- Department of Biological Sciences, National University of Singapore, Singapore.
Organizational Affiliation: