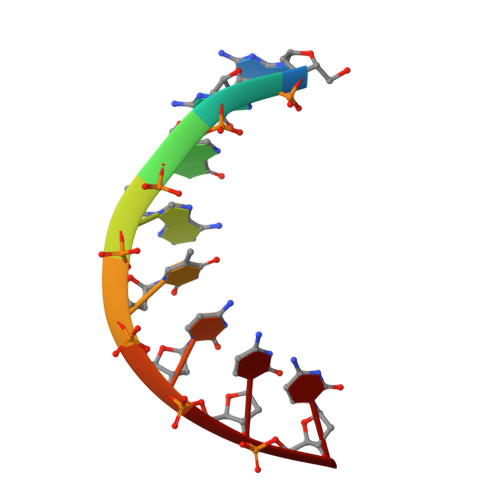
Three-dimensional structure of d(GGGATCCC) in the crystalline state.
Lauble, H., Frank, R., Blocker, H., Heinemann, U.(1988) Nucleic Acids Res 16: 7799-7816
- PubMed: 3166518
- DOI: https://doi.org/10.1093/nar/16.16.7799
- Primary Citation of Related Structures:
3ANA - PubMed Abstract:
The structure of the self-complementary octamer d(GGGATCCC) has been analysed by single crystal X-ray diffraction methods at a nominal resolution of 2.5 A. With acceptable stereochemistry of the model the crystallographic R factor was 16.6% after restrained least-squares refinement. In the crystal, d(GGGATCCC) forms an A-DNA double helix with slightly varying conformation of the two strands. The average displacement of the base pairs from the helix axis is unusually large and is accompanied by pronounced sliding of the base pairs along their long axes at all dinucleotide steps except for the central AT. With 12 base pairs per complete turn the helix is considerably underwound. As observed with most oligodeoxyribonucleotides analysed by X-ray crystallography so far, the octamer displays reduced base pair tilt, increased rise per base pair and a more open major groove compared with canonical A-DNA. We propose that, based on these parameters, three A-helical sub-families may be defined; d(GGGATCCC) then is a representative of the class with intermediate tilt, rise, and major groove width.
- Abteilung Saenger, Institut für Kristallographie, Freie Universität Berlin, FRG.
Organizational Affiliation: